Abstract
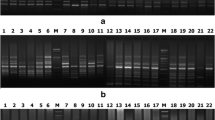
Rice is believed to have originated from Indo-China, area between China and India, and then spread throughout the world. The Indochina region mainly includes countries like Thailand, Laos and Vietnam, which are the world’s major rice exporters. Rice varieties grown in this area are highly diverse due to their different environment, ecosystem and climatic conditions. The objective of this study was to evaluate the genetic relationship of Indochina rice varieties using intersimple sequence repeat (ISSR), sequence-related amplified polymorphism (SRAP) and insertion–deletion (InDel) markers. Forty-six rice varieties, including 16, 4, 11 and 15 from Thailand, China, Laos and Vietnam, respectively were used in this study. Seventeen of the 20 ISSR primers showed 82.96% polymorphism. At the same time, 17 of the 30 primer pairs of SRAP marker showed clear DNA amplification, which resulted in 84.79% polymorphism. Ninety-seven of 133 InDel markers have about 99.47% polymorphism. Three markers showed average PIC score ranging from 0.20 to 0.26. When the analysis was conducted using UPGMA clustering method, it was found that the combined data from three markers gave a better result than each marker separately. The results from clustering analysis showed that all accessions can be grouped based on their location and can be categorized into two major groups. Useful results from this study could bring substantial benefits and ultimately help the rice breeders to develop elite rice varieties in future.




Similar content being viewed by others
References
Anderson J. A., Churchill G. A., Autrique J. E., Tanksley S. D. and Sorrells M. E. 1993 Optimizing parental selection for genetic linkage maps. Genome 36, 181–186.
Barrett B. A., Kidwell K. K. and Fox P. N. 1998 Comparison of AFLP and pedigree based genetic diversity assessment methods using wheat cultivars from the Pacific Northwest. Crop Sci. 38, 1271–1278.
Bhattramakki D., Dolan M., Hanafey M., Wineland R., Vaske D., Register I. J. C. et al. 2002 Insertion-deletion polymorphisms in 3’ regions of maize genes occur frequently and can be used as highly informative genetic markers. Plant Mol. Biol. 48, 539–547.
Bolaric S., Barth S., Melchinger A. E. and Posselt U. K. 2005 Genetic diversity in European perennial ryegrass cultivars investigated with RAPD markers. Plant Breed. 124, 161–166.
Britten R. J., Rowen L., Williams J. and Cameron R. A. 2003 Majority of divergence between closely related DNA samples is due to indels. Proc. Natl. Acad. Sci. USA 100, 4661–4665.
Budak H., Shearman R., Parmaksiz I. and Dweikat I. 2004 Comparative analysis of seeded and vegetative biotype buffalo-grasses based on phylogenetic relationship using ISSRs, SSRs, RAPDs, and SRAPs. Theor. Appl. Genet. 109, 280–288.
Chen S. Y., Dai T. X., Chang Y. T., Wang S. S., Ou S. L., Chuang W. L. et al. 2013 Genetic diversity among Ocimum species based on ISSR, RAPD and SRAP markers. Australian J. Crop Sci. 7, 1463–1471.
Choudhury B., Khan M. L. and Dayanandan S. 2013 Genetic structure and diversity of indigenous rice (Oryza sativa) varieties in the Eastern Himalayan region of Northeast India. Springer Plus 2, 228.
Doyle J. J. and Doyle J. L. 1987 A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Fernández M., Figueiras A. and Benito C. 2002 The use of ISSR and RAPD markers for detecting DNA polymorphism, genotype identification and genetic diversity among barley cultivars with known origin. Theor. Appl. Genet. 104, 845–851.
Fu L., Zhang H., Wu X., Li H., Wei H., Wu Q. et al. 2010 Evaluation of genetic diversity in Lentinulaedodes strains using RAPD, ISSR and SRAP markers. World J. Microbiol. Biotech. 26, 709–716.
Fu X., Ning G., Gao L. and Bao M. 2008 Genetic diversity of Dianthus accessions as assessed using two molecular marker systems (SRAPs and ISSRs) and morphological traits. Sci. Hort. 117, 263–270.
Fufa H., Baenziger P. S., Beecher I., Dweikat V., Graybosch R. A. and Eskridge K. M. 2005 Comparison of phenotypic and molecular marker-based classifications of hard red winter wheat cultivars. Euphytica 145, 133–146.
Fuller D. Q. 2011 Pathways to Asian civilizations: tracing the origins and spread of rice and rice cultures. Rice 4, 78–92.
García-Lor A., Luro F., Navarro L. and Ollitrault P. 2012 Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity: a perspective for genetic association studies. Mol. Genet. Genomics 287, 77–94.
Gross B. L. and Zhao Z. 2014 Archaeological and genetic insights into the origins of domesticated rice. Proc. Natl. Acad. Sci. USA 111, 6190–6197.
Hou X., Li L., Peng Z., Wei B., Tang S., Ding M. et al. 2010 A platform of high-density INDEL/CAPS markers for map-based cloning in Arabidopsis. Plant J. 63, 880–888.
Innark P., Ratanachan T., Khanobdee C., Samipak S. and Jantasuriyarat C. 2014 Downy mildew resistant/susceptible cucumber germplasm (Cucumis sativus L.) genetic diversity assessment using ISSR markers. Crop Prot. 60, 56–61.
Keyes C. F. 1995 The golden peninsula: culture and adaptation in mainland Southeast Asia. University of Hawaii Press, Honolulu.
Kladmook M., Kumchoo T. and Hongtraku V. 2012 Genetic diversity analysis and subspecies classification of Thailand rice landraces using DNA markers. Afr. J. Biotech. 11, 14044–14053.
Kojima T., Nagaoka T., Noda K. and Ogihara Y. 1998 Genetic linkage map of ISSR and RAPD markers in Einkorn wheat in relation to that of RFLP markers. Theor. Appl. Genet. 96, 37–45.
Li G. and Quiros C. F. 2001 Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor. Appl. Genet. 103, 455–461.
Li G., McVetty P. B. E. and Quiros C. F. 2013 SRAP molecular marker technology in plant science. In Plant breeding from laboratories to fields. (ed. B. Andersen). InTech, Copenhagen.
Liu J., Li J., Qu J. and Yan S. 2015 Development of genome-wide insertion and deletion polymorphism markers from next-generation sequencing data in rice. Rice 8, 63.
Mohanty S., Panda M. K., Acharya L. and Nayak S. 2014 Genetic diversity and gene differentiation among ten species of Zingiberaceae from Eastern India. 3 Biotech 4, 383–390.
Moonsap P., Laksanavilat N., Tasanasuwan P., Kate-Ngam S. and Jantasuriyarat C. 2019 Assessment of genetic variation of 15 Thai elite rice cultivars using InDel markers. Crop Breed. Appl. Biotech. 19, 15–21.
Ollitrault P., Terol J., Garcia-Lor A., Bérard A., Chauveau A., Froelicher Y. et al. 2012 SNP mining in C. clementina BAC end sequences; transferability in the Citrus genus (Rutaceae), phylogenetic inferences and perspectives for genetic mapping. BMC Genom. 13, 13.
Osorio C. E., Udall J. A., Salvo-Garrido H. and Maureira-Butler I. J. 2018 Development and characterization of InDel markers for Lupinusluteus L. (Fabaceae) and cross-species amplification in other Lupin species. Electron. J. Biotechnol. 31, 44–47.
Pakseresht F., Talebi R. and Karami E. 2013 Comparative assessment of ISSR, DAMD and SCoT markers for evaluation of genetic diversity and conservation of landrace chickpea (Cicer arietinum L.) genotypes collected from north-west of Iran. Physiol. Mol. Biol. Plants 19, 563–574.
Parsons B. J., Newbury H. J., Jackson M. T. and Ford-Lloyd B. V. 1997 Contrasting genetic diversity relationships are revealed in rice (Oryza sativa L.) using different marker types. Mol. Breed. 3, 115–125.
Patel S., Ravikiran R., Chakraborty S., Macwana S., Sasidharan N., Trivedi R. et al. 2014 Genetic diversity analysis of colored and white rice genotypes using microsatellite (SSR) and insertion-deletion (INDEL) markers. Emir. J. Food Agri. 26, 497.
Peakall R. and Smouse P. E. 2012 GenAlEx 6.5: Genetic analysis in excel. Population Genetic Software for Teaching and Research-An Update. Bioinformatics 28, 2537–2539.
Prevost A. and Wilkinson M. J. 1999 A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor. Appl. Genet. 98, 107–112.
Raman H., Raman R., Wood R. and Martin P. 2006 Repetitive Indel markers within the ALMT1 gene conditioning aluminium tolerance in wheat (Triticum aestivum L.). Mol. Breed. 18, 171–183.
Rohlf F. J. 2000 NTSYSpc: Numerical taxonomy and multivariate analysis system. Version 2.02, Exeter Software, Setauket, New York.
Roldán-Ruiz I., Dendauw J., Van-Bockstaele E., Depicker A. and De-Loose M. 2000 AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol. Breed. 6, 125–134.
Sneath P. H. and Sokal R. R. 1973 Numerical Taxonomy: the principles and practice of numerical classification. 1st Edition, W. H. Freeman, San Francisco.
Subudhi P. K., De Leon T. B., Tapia R., Chai C., Karan R., Ontoy J. et al. 2018 Genetic interaction involving photoperiod-responsive hd1 promotes early flowering under long-day conditions in rice. Sci. Rep. 8, 2081.
USDA Office of Global Analysis. 2017 Grain: World markets and trade, 08.10.2017. Department of Agriculture, IOP Publishing, United States (http://usda.mannlib.cornell.edu/usda/fas/grain-market//2010s/2017/grain-market-08-10-2017.pdf), accessed 20 November 2017.
Wanchana S., Toojinda T., Tragoonrung S. and Vanavichit A. 2003 Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.). Plant Sci. 165, 1193–1199.
Wu D. H., Wu H. P., Wang C. S., Tseng H. Y. and Hwu K. K. 2013 Genome-wide indel marker system for application in rice breeding and mapping studies. Euphytica 192, 131–143.
Yano M., Kojima S., Takahashi Y., Lin H. and Sasaki T. 2001 Genetic control of flowering time in rice, a short-day plant. Plant Physiol. 127, 1425–1429.
Yap I. V. and Nelson R. J. 1996 WINBOOT: a program for performing bootstrap analysis for binary data to determine the confidence limits of UPGMA-based dendrograms. In IRRI Disc. Ser. No. 14. Int. Rice Res. Inst., Manila, Philippines.
Yeh F. C. and Yang R. C. 1999 Popgene version 1.32 Microsoft Window-based freeware for Population Genetic Analysis, Quick User Guide. University of Alberta And Tim Boyle, Centre for International Forestry Research (http://www.ualberta.ca/~fyeh/fyeh/).
Zietkiewicz E., Rafalski A. and Labuda D. 1994 Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20, 176–183.
Acknowledgements
This research was supported from graduate scholarship provided by the National Research Council of Thailand (NRCT) in the 2018 fiscal year and the Graduate Scholarship from the Graduate School of Kasetsart University and the Bilateral Research Co-operation, Faculty of Science, Kasetsart University to Miss Pattaraborn Moonsap.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: H. A. Ranganath
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Moonsap, P., Laksanavilat, N., Sinumporn, S. et al. Genetic diversity of Indo-China rice varieties using ISSR, SRAP and InDel markers. J Genet 98, 80 (2019). https://doi.org/10.1007/s12041-019-1123-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-019-1123-0