Abstract
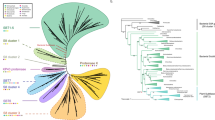
Despite the ubiquitous occurrence of heat-shock protein 60 (Hsp60) and their role in maintenance of cell activity and integrity, this protein remains poorly characterized in many of the symbiotic soil mycorrhizal fungi such as Rhizophagus irregularis. Thus, in the current study, an attempt has been made to elucidate the evolutionary history, time of divergence followed by estimation of population genetic parameters of hsp60 using R. irregularis as a model organism. Sequence alignment reported here identified several close homologues for hsp60 (gene) and Hsp60 (protein) from diverse taxa, while the output from protein-based phylogenetic tree indicates that mitochondrial Hsp60 of R. irregularis shares close evolutionary relationship with classical \(\alpha \)-proteobacteria. This is perhaps the first line of evidence elucidating the likelihood of hsp60 from fungal taxa sharing a close evolutionary relationship with classical \(\alpha \)-proteobacteria as a common ancestor. Comprehensive analysis of mitochondrial hsp60 from selected fungal taxa from the evolutionary point of view explains the possibility of gene duplication and or horizontal gene transfer of this gene across various fungal species. Synteny relationships and population genetics credibly explain high genetic variability associated with fungal hsp60 presumably brought by random genetic recombination events. The results presented here also confirm a high level of genetic differentiation of hsp60 among all the three fungal populations analysed. In this context, the outcome of the current study, based on computational approach, stands as a testimony for explaining the possibility of increased genetic differentiation experienced by hsp60 of R. irregularis.






Similar content being viewed by others
References
Aguileta G., Refregier G., Yockteng R., Fournier E. and Giraud T. 2009 Rapidly evolving genes in pathogens: methods for detecting positive selection and examples among fungi, bacteria, viruses and protists. Infect. Genet. Evol. 9, 656–670.
Alba-Fierro C. A., Pérez-Torres A., Toriello C., Pulido-Camarillo E., López-Romero E., Romo-Lozano Y. et al. 2016 Immune response induced by an immunodominant 60 kDa glycoprotein of the cell wall of Sporothrix schenckii in two mice strains with experimental sporotrichosis. J. Immunol. Res. 2016, article ID 6525831.
Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W. et al. 1997 Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Anderson J. B., Kohn L. M. and Leslie J. F. 1992 Genetic mechanisms in fungal adaptation. The fungal community: Its organization and role in the ecosystem, 2nd edition, pp. 73–98. Marcel Dekker, New York.
Beaudet D., de la Providencia I. E., Labridy M., Roy-Bolduc A., Daubois L. and Hijri M 2014 Intraisolate mitochondrial genetic polymorphism and gene variants coexpression in arbuscular mycorrhizal fungi. Genome Biol. Evol. 7, 218–227.
Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G. et al. 2013 MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319.
Blaiotta G., Fusco V., Ercolini D., Aponte M., Pepe O. and Villani F. 2008 Lactobacillus strain diversity based on partial hsp60 gene sequences and design of PCR-restriction fragment length polymorphism assays for species identification and differentiation. Appl. Environ. Microbiol. 74, 208–215.
Boisvert D. C., Wang J., Otwinowski Z., Norwich A. L. and Sigler P. B. 1996 The 2.4 Å crystal structure of the bacterial chaperonin GroEL complexed with ATP\(\gamma \)S. Nat. Struct. Mol. Biol. 3, 170.
Boon E., Zimmerman E., St-Arnaud M. and Hijri M. 2013 Allelic differences within and among sister spores of the arbuscular mycorrhizal fungus Glomus etunicatum suggest segregation at sporulation. PLoS One 8, 8330.
Borchiellini C., Boury-Esnault N., Vacelet J. and Le Parco Y. 1998 Phylogenetic analysis of the Hsp70 sequences reveals the monophyly of Metazoa and specific phylogenetic relationships between animals and fungi. Mol. Biol. Evol. 15, 647–655.
Borneman A. R., Forgan A. H., Kolouchova R., Fraser J. A. and Schmidt S. A. 2016 Whole genome comparison reveals high levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae. G3: Genes, Genomes, Genetics (Bethesda) 6, 957–971.
Börstler B., Raab P. A., Thiéry O., Morton J. and Redecker D. 2008 Genetic diversity of the arbuscular mycorrhizal fungus Glomus intraradices as determined by mitochondrial large subunit rRNA gene sequences is considerably higher than previously expected. New Phytol. 180, 452–465.
Brennwald A. and Redecker D. 2005 Mitochondrial large ribosomal subunit sequences are homogeneous within isolates of Glomus (arbuscular mycorrhizal fungi, Glomeromycota). Mycol. Res. 109, 1315–1322.
Brinig M. M., Cummings C. A., Sanden G. N., Stefanelli P., Lawrence A. and Relman D. A. 2006 Significant gene order and expression differences in Bordetella pertussis despite limited gene content variation. J. Bacteriol. 188, 2375–2382.
Brocchieri L. and Karlin S. 2000 Conservation among HSP60 sequences in relation to structure, function, and evolution. Protein Sci. 9, 476–486.
Bukau B. and Horwich A. L. 1998 The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366.
Campbell A. M. 2000 Lateral gene transfer in prokaryotes. Theor. Popul. Biol. 57, 71–77.
Cappello F., Conway de Macario E., Marasà L., Zummo G. and Macario A. J. 2008 Hsp60 expression, new locations, functions, and perspectives for cancer diagnosis and therapy. Cancer Biol. Ther. 7, 801–809.
Chakraborty U., Chakraborty B., Dey P. and Chakraborty A. P. 2015 Role of microorganisms in alleviation of abiotic stresses for sustainable agriculture. Abiotic stresses in crop plants, pp. 232–253. CABI, Wallingford.
Cheng M. Y., Hartl F. U., Martin J., Pollock R. A., Kalousek F., Neuper W. et al. 1989 Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 337, 620.
Cohen S. B. and Dor R. 2018 Phenotypic divergence despite low genetic differentiation in house sparrow populations. Sci. Rep. 8, 394.
Croll D., Wille L., Gamper H. A., Mathimaran N., Lammers P. J., Corradi N. et al. 2008 Genetic diversity and host plant preferences revealed by simple sequence repeat and mitochondrial markers in a population of the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 178, 672–687.
De la Cruz F. and Davies J. 2000 Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 8, 128–133.
Drummond A. J., Rambaut A., Shapiro B. and Pybus O. G. 2005 Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192.
Drummond A. J., Suchard M. A., Xie D. and Rambaut A. 2012 Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973.
Durbin R., Eddy S. R., Krogh A. and Mitchison G. 1998 Biological sequence analysis: Probabilistic models of proteins and nucleic acids. Cambridge University Press, Cambridge UK.
Duret L. and Arndt P. F. 2008 The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 4, e1000071.
Ehinger M. O., Croll D., Koch A. M. and Sanders I. R. 2012 Significant genetic and phenotypic changes arising from clonal growth of a single spore of an arbuscular mycorrhizal fungus over multiple generations. New Phytol. 196, 853–861.
Excoffier L. and Lischer H. E. 2010 Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567.
Felsenstein J. 1981 Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Fitzpatrick D. A., Logue M. E., Stajich J. E. and Butler G. 2006 A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 6, 99.
Formey D., Molès M., Haouy A., Savelli B., Bouchez O., Bécard G. et al. 2012 Comparative analysis of mitochondrial genomes of Rhizophagus irregularis—syn. Glomus irregulare – reveals a polymorphism induced by variability generating elements. New Phytol. 196, 1217–1227.
Fu Y. X. 1997 Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925.
Gadkar V. and Rillig M. C. 2006 The arbuscular mycorrhizal fungal protein glomalin is a putative homolog of heat shock protein 60. FEMS Microbiol. Lett. 263, 93–101.
Galagan J. E., Henn M. R., Ma L. J., Cuomo C. A. and Birren B. 2005 Genomics of the fungal kingdom: insights into eukaryotic biology. Genome Res. 15, 1620–1631.
Galtier N., Nabholz B., Glémin S. and Hurst G. D. D. 2009 Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 18, 4541–4550.
Gupta R. S. 1995a Evolution of the chaperonin families (HSP60, HSP 10 and TCP-1) of proteins and the origin of eukaryotic cells. Mol. Microbiol. 15, 1–11.
Gupta R. S. 1995b Phylogenetic analysis of the 90 kD heat shock family of protein sequences and an examination of the relationship among animals, plants, and fungi species. Mol. Biol. Evol. 12, 1063–1073.
Gupta R. S. and Golding G. B. 1993 Evolution of HSP70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J. Mol. Evol. 37, 573–582.
Gupta S. and Knowlton A. A. 2007 HSP60 trafficking in adult cardiac myocytes: role of the exosomal pathway. Am. J. Physiol.-Heart Circ. Physiol. 292, H3052–H3056.
Hall T. A. 1999 Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98.
Hamrick J. L. and Godt M. W. 1990 Allozyme diversity in plant species. Plant population genetics, breeding, and genetic resources, pp. 43–63. CABI, USA.
Hartl D. L. and Clark A. G. 1997 Principles of population genetics, 4th edition, pp. 318–372. Sinauer Associates, Sunderland.
Hartl F. U. and Hayer-Hartl M. 2002 Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852–1858.
Heckman D. S., Geiser D. M., Eidell B. R., Stauffer R. L., Kardos N. L. and Hedges S. B. 2001 Molecular evidence for the early colonization of land by fungi and plants. Science 293, 1129–113.3.
Hedges S. B. and Kumar S. 2009 Discovering the timetree of life. The timetree of life, 3rd edition. Oxford, New York, pp. 3–18.
Heitman J. 2015 Evolution of sexual reproduction: a view from the fungal kingdom supports an evolutionary epoch with sex before sexes. Fungal. Biol. Rev. 29, 108–117.
Ho S. Y. 2007 Calibrating molecular estimates of substitution rates and divergence times in birds. J. Avian Biol. 38, 409–414.
Holbrook E. D. and Rappleye C. A. 2008 Histoplasma capsulatum pathogenesis: making a lifestyle switch. Curr. Opin. Microbiol. 11, 318–324.
Judson O. P. and Normark B. B. 1996 Ancient asexual scandals. Trends Ecol. Evol. 11, 41–46.
Karlin S. and Brocchieri L. 2000 Heat shock protein 60 sequence comparisons: duplications, lateral transfer, and mitochondrial evolution. Proc. Natl. Acad. Sci. USA 97, 11348–11353.
Kaufman B. A., Kolesar J. E., Perlman P. S. and Butow R. A. 2003 A function for the mitochondrial chaperonin Hsp60 in the structure and transmission of mitochondrial DNA nucleoids in Saccharomyces cerevisiae. J. Cell Biol. 163, 457–461.
Kuhn G., Hijri M. and Sanders I. R. 2001 Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature 414, 745.
Kumar S., Stecher G. and Tamura K. 2016 MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874.
Lamoth, F., Juvvadi, P. R., Soderblom, E. J., Moseley, M. A., Asfaw, Y. G. and Steinbach W. J. 2014 Identification of a key lysine residue in heat shock protein 90 required for azole and echinocandin resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 58, 1889–1896.
Lane N. and Martin W. 2010 The energetics of genome complexity. Nature 467, 929.
Lang B. F., Gray M. W. and Burger G. 1999 Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33, 351–397.
Latef A. A. H. A., Hashem A., Rasool S., Abd\_Allah E. F., Alqarawi A. A., Egamberdieva D. et al. 2016 Arbuscular mycorrhizal symbiosis and abiotic stress in plants: a review. J. Plant Biol. 59, 407–426.
Lee J. and Young J. P. W. 2009 The mitochondrial genome sequence of the arbuscular mycorrhizal fungus Glomus intraradices isolate 494 and implications for the phylogenetic placement of Glomus. New Phytol. 183, 200–211.
Leonhardt S. A., Fearson K., Danese P. N. and Mason T. L. 1993 HSP78 encodes a yeast mitochondrial heat shock protein in the Clp family of ATP-dependent proteases. Mol. Cell. Biol. 13, 6304–6313.
Lindquist S. and Craig E. A. 1988 The heat-shock proteins. Annu. Rev. Genet. 22, 631–677.
Liu W., Zhang Y., Jiang S., Deng Y., Christie P., Murray P. J. et al. 2016 Arbuscular mycorrhizal fungi in soil and roots respond differently to phosphorus inputs in an intensively managed calcareous agricultural soil. Sci. Rep. 6, 24902.
López-García P. and Moreira D. 1999 Metabolic symbiosis at the origin of eukaryotes. Trends Biochem. Sci. 24, 88–93.
Lorch M., Mason J. M., Sessions R. B. and Clarke A. R. 2000 Effects of mutations on the thermodynamics of a protein folding reaction: implications for the mechanism of formation of the intermediate and transition states. Biochemistry 39, 3480–3485.
Lucking R., Huhndorf S., Pfister D. H., Plata E. R. and Lumbsch H. T. 2009 Fungi evolved right on track. Mycologia 101, 810–822.
Marleau J., Dalpé Y., St-Arnaud M. and Hijri M. 2011 Spore development and nuclear inheritance in arbuscular mycorrhizal fungi. BMC Evol. Biol. 11, 51.
McDonald B. A. 1997 The population genetics of fungi: tools and techniques. Phytopathology 87, 448–453.
Meng Q., Li B. X. and Xiao X. 2018 Toward developing chemical modulators of Hsp60 as potential therapeutics. Front. Mol. Biosci. 5, 35.
Muse S. V. and Gaut B. S. 1994 A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 11, 715–724.
Naylor D. J. and Hartl F. U 2001 Contribution of molecular chaperones to protein folding in the cytoplasm of prokaryotic and eukaryotic cells. Biochem. Soc. Symp. 68, 45–68.
Nei M. and Kumar S. 2000 Molecular evolution and phylogenetics. Oxford, New York.
Nybom H. and Bartish I. V. 2000 Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect. Plant. Ecol. Evol. Syst. 3, 93–114.
Ostermann J., Horwich A. L., Neupert W. and Hartl F. U. 1989 Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature 341, 125.
Pond S. L. K. and Muse S. V. 2005 Hyphy: Hypothesis testing using phylogenies. Statistical methods in molecular evolution, pp. 125–181. Springer, New York.
Porcel R., Aroca R., Cano C., Bago A. and Ruiz-Lozano J. M. 2006 Identification of a gene from the arbuscular mycorrhizal fungus Glomus intraradices encoding for a 14-3-3 protein that is up-regulated by drought stress during the AM symbiosis. Microb. Ecol. 52, 575.
Priya S., Sharma S. K. and Goloubinoff P. 2013 Molecular chaperones as enzymes that catalytically unfold misfolded polypeptides. FEBS Lett. 587, 1981–1987.
Raggam R. B., Salzer H. J., Marth E., Heiling B. and Paulitsch A. H. and Buzina W. 2011 Molecular detection and characterisation of fungal heat shock protein 60. Mycoses 54, e394–e399.
Rambaut A. and Drummond A. J. 2010 Treeannotator version 1.6. 1. University of Edinburgh, Edinburgh, UK.
Redecker D., Morton J. B. and Bruns T. D. 2000 Ancestral lineages of arbuscular mycorrhizal fungi (Glomales). Mol. Phylogenet. Evol. 14, 276–284.
Richards T. A., Soanes D. M., Foster P. G., Leonard G., Thornton C. R. and Talbot N. J. 2009 Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell. 21, 1897–1911.
Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J. C., Guirao-Rico S., Librado P., Ramos-Onsins S. E. et al. 2017 DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302.
Schüßler A., Schwarzott D and Walker C. 2001 A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105, 1413–1421.
Sharma M., Fomda B. A., Mazta S., Sehgal R., Singh B. B. and Malla N. 2013 Genetic diversity and population genetic structure analysis of Echinococcus granulosus sensu stricto complex based on mitochondrial DNA signature. PLoS One 8, e82904.
Smith S. E. and Read D. J. 2010 Mycorrhizal symbiosis. Academic Press, New York.
Soltys B. J. and Gupta R. S. 1997 Cell surface localization of the 60 kDa heat shock chaperonin protein (hsp60) in mammalian cells. Cell Biol. Int. 21, 315–320.
Somel M., Wilson Sayres M. A., Jordan G., Huerta-Sanchez E., Fumagalli M., Ferrer-Admetlla A. et al. 2013 A scan for human-specific relaxation of negative selection reveals unexpected polymorphism in proteasome genes. Mol. Biol. Evol. 30, 1808–1815.
Suzuki Y. and Gojobori T. 1999 A method for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 16, 1315–1328.
Tajima F. 1989 Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
Tajima F. 1993 Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135, 599–607.
Tamura K. and Nei M. 1993 Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526.
Thompson J. D., Higgins D. G. and Gibson T. J. 1994 CLUSTAL w: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Tisserant E., Malbreil M., Kuo A., Kohler A., Symeonidi A., Balestrini R. et al. 2013 Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. USA 110, 20117–20122.
Tiwari S., Thakur R. and Shankar J. 2015 Role of heat-shock proteins in cellular function and in the biology of fungi. Biotechnol. Res. Int. (https://doi.org/10.1155/2015/132635).
Trent J. D. 1996 A review of acquired thermotolerance, heat-shock proteins, and molecular chaperones in archaea. FEMS Microbiol. Rev. 18, 249–258.
Xiong J., Rayner S., Luo K., Li Y. and Chen S. 2006 Genome wide prediction of protein function via a generic knowledge discovery approach based on evidence integration. BMC Bioinf. 7, 268.
Xu Z., Horwich A. L. and Sigler P. B. 1997 The crystal structure of the asymmetric GroEL–GroES–(ADP) 7 chaperonin complex. Nature 388, 741.
Yang Z. and Yoder A. D. 1999 Estimation of the transition/transversion rate bias and species sampling. J. Mol. Evol. 48, 274–283.
Yang Z. and Nielsen R. 2000 Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17, 32–43.
Acknowledgements
We acknowledge Jain University for providing the facilities necessary to carry out our work and for providing the University PhD research scholarship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding Editor: Manoj Prasad
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mothay, D., Ramesh, K.V. Evolutionary history and genetic diversity study of heat-shock protein 60 of Rhizophagus irregularis. J Genet 98, 48 (2019). https://doi.org/10.1007/s12041-019-1096-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-019-1096-z