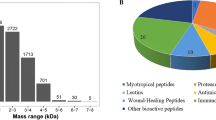
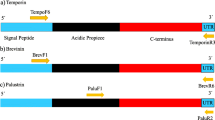
Abstract
Amphibian secretion is an important source of bioactive molecules that naturally protect the skin against noxious microorganisms. Collectively called antimicrobial peptides (AMPs), these molecules have a wide spectrum of action, targeting viruses, bacteria and fungi. Like many membrane and secreted proteins, AMPs have cleavable signal sequences that mediate and translocate the nascent polypeptide chains into the endoplasmic reticulum. Although it is accepted that the signal peptides (SPs) are simple and interchangeable, there is neither sequence nor structure that is conserved among all gene families. They derived from a common ancestor but developed different traits as they adapt to distinct environmental pressures. The aim of this study was to provide an overview of the diversity of SPs of the frog, taking into account reported cDNA sequences and the evolutionary relationship among them. We analysed more than 2000 records that reported the relative abundance, diversity and evolutionary divergence based on the peptide signals of frog AMPs. We conclude that the physical properties of the sequence are more important than the specific peptides in AMP SPs. Since there is significant overlapping among related genera, differences in secretion from different peptide types should be regulated by additional levels, such as posttranscriptional modifications or 5-UTR sequences.




Similar content being viewed by others
References
Amiche M., Seon A. A., Pierre T. N. and Nicolas P. 1999 The dermaseptin precursors: a protein family with a common preproregion and a variable C-terminal antimicrobial domain. FEBS Lett. 456, 352–356.
Brahmachary M., Krishnan S. P. T., Koh J. L. Y., Khan A. M., Seah S. H., Tan T. W. et al. 2004 ANTIMIC: a database of antimicrobial sequences. Nucleic Acids Res. 32, D586–D589.
Conlon J. M. 2008 Reflections on a systematic nomenclature for antimicrobial peptides from the skins of frogs of the family Ranidae. Peptides 29, 1815–1819.
Csordas A. and Michl H. 1969 Primary structure of two oligopeptides of the toxin of Bombina variegate L. Toxicon 7, 103–108.
Edgar R. C. 2004 MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Excoffier L., Smouse P. E. and Quattro J. M. 1992 Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491.
Fjell C. D., Hancock R. E. W. and Cherkasov A. 2007 AMPer: a database and an automated discovery tool for antimicrobial peptides. Bioinformatics 23, 1148–1155.
Gallo R. L. and Huttner K. M. 1998 Antimicrobial peptides: an emerging concept in cutaneous biology. J. Invest. Dermatol. 111, 739–743.
Guo C., Hu Y., Li J., Liu Y., Li S. et al. 2014 Identification of multiple peptides with antioxidant and antimicrobial activities from the skin and its secretions of Hylarana taipehensis, Amolops lifanensis, and Amolops granulosus. Biochimie 105, 192–201.
Gupta G. K., Joshi T., Madhudiya K., Biswas K., Bhatanagar S., Ranawat B. et al. 2017 Antibacterial activity of skin secretion of toad Bufo melanostictus and frog Rana cyanophylictitus. Int. J. Acad. Res. Dev. 2, 684–686.
Hon L. S., Zhang Y., Kaminker J. S. and Zhang Z. 2009 Computational prediction of the functional effects of amino acid substitutions in signal peptides using a model-based approach. Hum. Mutat. 30, 99–106.
Kamvar Z. N., Tabima J. F. and Grünwald N. J. 2014 Poppr: an R package for genetic analysis of populations with clonal, parti ally clonal, and/or sexual reproduction. PeerJ 2, e281.
König E. and Bininda-Emonds O. R. P. 2011 Evidence for convergent evolution in the antimicrobial peptide system in anuran amphibians. Peptides 32, 20–25.
König E., Bininda-Emonds O. R. P. and Shaw C. 2015 The diversity and evolution of anuran skin peptides. Peptides 63, 96–117.
Lodish H., Berk A., Zipursky S. L., Matsudaira P., Baltimore D. and Darnell J. 2000 Molecular cell biology, 4th edition. W. H. Freeman & Company, New York.
Maselli V. M., Bilusich D., Bowie J. H. and Tyler M. J. 2006 Host-defence skin peptides of the Australian Streambank Froglet Crinia riparia: isolation and sequence determination by positive and negative ion electrospray mass spectrometry. Rapid. Commun. Mass Spectrom. 20, 797–803.
Mechkarska M., Prajeep M., Leprince J., Vaudry H., Meetani M. A., Evans B. J. and Conlon J. M. 2013 A comparison of host-defense peptides in skin secretions of female Xenopus laevis \(\times \) Xenopus borealis and X. borealis \(\times \) X. laevis F1 hybrids. Peptides 45, 1–8.
Miller K. W., Evans E. J., Eisenberg S. P. and Thompson R. C. 1989 Secretory leukocyte protease inhibitor binding to mRNA and DNA as a possible cause of toxicity to Escherichia coli. J. Bacteriol. 171, 2166–2172.
Miller M. A., Pfeiffer W. and Schwartz T. 2010 Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans.
Morris E. K., Caruso T., Buscot F., Fischer M., Hancock C., Maier T. S. et al. 2014 Choosing and using diversity indices: insights for ecological applications from the German Biodiversity Exploratories. Ecol. Evol. 4, 3514–3524.
Nicolas P., Vanhoye D. and Amiche M. 2003 Molecular strategies in biological evolution of antimicrobial peptides. Peptides 24, 1669–1680.
Olivera B. M. and Teichert R. W. 2007 Diversity of the neurotoxic Conus peptides: a model for concerted pharmacological discovery. Mol. Interventions 7, 251–260.
Pagès H., Aboyoun P., Gentleman R. and DebRoy S. 2017. Biostrings: efficient manipulation of biological strings. R package version 2.46.0.
Park C. B., Kim M. S. and Kim S. C. 1996 A novel antimicrobial peptide from Bufo bufo gargarizans. Biochem. Biophys. Res. Commun. 218, 408–413.
Petersen N. T., Brunak S., von Heijne G. and Nielsen H 2011 SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786.
Pyron R. A. and Wiens J. J. A. 2011 Large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 61, 543–583.
Simmaco M., Kreil G. and Barra D. 2009 Bombinins, antimicrobial peptides from Bombina species. Biochim. Biophys. Acta 1788, 1551–1555.
Simmaco M., Mignona G., Barra D. and Bossa F. 1994 Antimicrobial peptides from skin secretions of Rana esculenta - molecular cloning of cDNA encoding and isolation of new active peptides. J. Biol. Chem. 269, 11956–11961.
Smith A. B., Daly N. L. and Craik D. J. 2011 Cyclotides: a patent review. Expert Opin. Ther. Pat. 21, 1657–1672.
Tang X., Zhang Y., Hu W., Xu D., Tao H., Yang X. et al. 2010 Molecular diversification of peptide toxins from the tarantula Haplopelma hainanum (Ornithoctonus hainana) venom based on transcriptomic, peptidomic, and genomic analyses. J. Proteome Res. 9, 2550–2564.
Uhlig T., Kyprianou T., Martinelli F. G., Oppici C. A., Heiligers D., Hills D. R. J. et al. 2014 The emergence of peptides in the pharmaceutical business: from exploration to exploitation. EuPA Open Proteomics 4, 58–69.
Wang G., Li X. and Wang Z. 2016 APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093.
Wang Z. and Wang G. 2004 APD: the antimicrobial peptide database. Nucleic Acids Res. 32, D590–D592.
Wu D., Gao Y., Qi Y., Chen L., Ma Y. and Li Y. 2014 Peptide-based cancer therapy: opportunity and challenge. Cancer Lett. 351, 13–22.
Acknowledgements
This work was supported by grants from the National Council for Scientific and Technological Research (CONICET) (PIP no. 11220120100050OC). This research is framed within the P-UE CONICET no. 22920160100044. NLC is grateful to CONICET for her grant. LOP and MMM are researchers of the CONICET. The authors state that there is no conflict of interest regarding the contents of this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: H. A. Ranganath
Rights and permissions
About this article
Cite this article
Pérez, L.O., Cancelarich, N.L., Aguilar, S. et al. Genetic analysis of signal peptides in amphibian antimicrobial secretions. J Genet 97, 1205–1212 (2018). https://doi.org/10.1007/s12041-018-1018-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-018-1018-5