Abstract
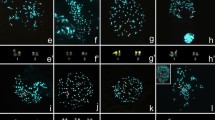
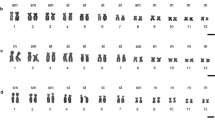
We conducted a cytogenetic study of four hyline frog species (Dendropsophus elegans, D. microps, D. minutus and D. werneri) from southern Brazil. All species had 2n = 30 chromosomes, with interspecific and intraspecific variation in the numbers of metacentric, submetacentric, subtelocentric and telocentric chromosomes. C-banding and fluorochrome staining revealed conservative GC-rich heterochromatin localized in the pericentromeric regions of all species. The location of the nucleolus organizer regions, as confirmed by fluorescent in situ hybridization, differed between species. Telomeric probes detected sites that were restricted to the terminal regions of all chromosomes and no interstitial or centromeric signals were observed. Our study corroborates the generic synapomorphy of 2n = 30 chromosomes for Dendropsophus and adds data that may become useful for future taxonomic revisions and a broader understanding of chromosomal evolution among hylids.





Similar content being viewed by others
References
Amaro-Ghilardi R. C., Silva M. J. J., Rodrigues M. T. and Yonenaga-Yassuda Y. 2008 Chromosomal studies in four species of genus Chaunus (Bufonidae, Anura): localization of telomeric and ribosomal sequences after fluorescence in situ hybridization (FISH). Genetica 134, 159–168.
Badissera Jr F. A., Oliveira P. S. L. and Kasahara S. 1993 Cytogenetics of four Brazilian Hyla species (Amphibia, Anura) and description of a case of supernumerary chromosome. Rev. Brasil Gen. 16, 335–345.
Bogart J. 1973 Evolution of anuran karyotypes. In: Evolutionary biology of Anurans (ed. J. Vial). University of Missouri Press, Columbia.
Campos J. R. C., Ananaias F., Brasileiro C., Yamamoto M., Haddad C. F. B. and Kasahara S. 2009 Chromosome evolution in three Brazilian Leptodactylus species (Anura, Leptodactylidae), with phylogenetic considerations. Hereditas 146, 104–111.
Catroli G. F. and Kasahara S. 2009 Cytogenetic data on species of the family Hylidae (Amphibia, Anura): results and perspectives. Publ. UEPG CiBiol. Saúde 15, 67–86.
Carvalho K. A., Garcia P. C. and Recco-Pimentel S. M. 2009 NOR dispersion, telomeric sequence detection in centromeric regions and meiotic multivalent configurations in species of the Aplastodiscus albofrenatus group (Anura, Hylidae). Cytogenet. Genome Res. 126, 359–367.
Datson P. M. and Murray B. G. 2006 Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism? Chromosome Res. 14, 845–857.
Faivovich J., Haddad C. F. B., Garcia P. C. A., Frost D. R., Campbell, J. A. and Wheeler W. D. 2005 A systematics review of the frog family Hylidae, with special reference to the Hylinae, a phylogenetic analysis and taxonomic revision. Bull. Am. Mus. Nat. Hist. 294, 1–240.
Fouquet A., Noona B. P., Blanc M. and Orrico V. G. D. 2011 New distributional records and phylogenetic position for Dendropsophus gaucheri. Zootaxa 3035, 59–67.
Frost D. R. 2014 Amphibian species of the world: an online reference, version 6.0 (accessed on 13th March 2014). Electronic database accessible at http://research.amnh.org/herpetology/amphibia/index.html.
Green D. M. and Sessions S. K. 1991 Nomenclature for chromosomes. In Amphibian cytogenetics and evolution (ed. D. M. Green and S. K. Sessions). Academic Press, San Diego.
Gruber S. L., Haddad C. F. B. and Kasahara S. 2005 Evaluating the karyotypic diversity in species of Hyla (Anura; Hylidae) with 2n = 30 chromosomes based on the analysis of ten species. Folia Biol. 51, 68–75.
Hatanaka T. and Galetti P. M. 2004 Mapping of the 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus Agassiz, 1829 (Characiformes, Prochilodontidae). Genetica 122, 239–244.
Howell W. M. and Black D. A. 1980 Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer. Experientia 36, 1014–1015.
Ijdo J. W., Wells R. A., Baldini A. and Reeders S. T. 1991 Improved telomere detection using a telomere repeat probe (TTAGGG) generated by PCR. Nucleic Acids Res. 19, 4780.
IUCN 2013 The IUCN red list of threatened species, version 2013.2 (accessed on 2nd February 2013). Electronic database accessible at http://www.iucnredlist.org.
Kaiser H., Mais C., Bolaños F., Steinlein C., Feichtinger W. and Schmid M. 1996 Chromosomal investigation of three Costa Rican frogs from the 30-chromosome radiation of Hyla with the description of a unique geographic variation in nucleolus organizer regions. Genetica 98, 95–102.
Kellogg R. 1932 Mexican tailless amphibians in the United States National Museum. Bull. U. S. Natl. Mus. 160, 1–224.
King M., Contreras N. and Honeycutt R. L. 1990 Variation within and between nucleolar organizer regions in Australian hylid frogs (Anura) shown by 18S + 28S in-situ hybridization. Genetica 80, 17–29.
Medeiros L. R., Rossa-Feres D. C. and Recco-Pimentel S. M. 2003 Chromosomal differentiation of Hyla nana and Hyla sanborni (Anura, Hylidae) with a description of NOR polymorphism in H. nana. J. Hered. 94, 149–154.
Medeiros L. R., Rossa-Feres D. C., Jim J. and Recco-Pimentel S. M. 2006 B-chromosomes in two Brazilian populations of Dendropsophus nanus (Anura, Hylidae). Genet. Mol. Biol. 29, 257–262.
Medeiros L. R., Lourenço L. B., Rossa-Feres D. C., Lima A. P., Andrade G. V., Giaretta A. et al. 2013 Comparative cytogenetic analysis of some species of the Dendropsophus microcephalus group (Anura, Hylidae) in the light of phylogenetic inferences. BMC Genet. 14, 59–76.
Noleto R. B., Amaro R. C., Verdade V. K., Campos J. R. C., Gallego L. F. K., Lima A. M. X. et al. 2011 Comparative cytogenetics of eight species of Cycloramphus (Anura, Cycloramphidae). Zool. Anz. 250, 205–214.
Pyron R. A. and Wiens J. J. 2011 A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 61, 543–583.
Rocchi M., Archidiacono N., Schempp W., Capozzi O. and Stanyon R. 2012 Centromere repositioning in mammals. Heredity 108, 59–67.
Schmid M., Steinlein C., Bogart J. P., Feichtinger W., León P., La Marca E. et al. 2010 The chromosomes of terraranan frogs. Insights into vertebrate cytogenetics. Cytogenet. Genome Res. 130–131, 1–568.
Schweizer D. 1981 Counterstain-enhanced chromosome banding. Hum. Genet. 57, 1–14.
Skuk G. and Langone J. A. 1992 Los cromosomas de cuatroespeciesdel género Hyla (Anura: Hylidae) com número diploide de 2n = 30. Acta Zool. Lilloana 41, 165–171.
Suárez P., Cardozo D., Baldo D., Pereyra M. O., Faivovich J., Orrico V. G. D. et al. 2013 Chromosome evolution in Dendropsophini (Amphibia, Anura, Hylidae). Cytogenet. Genome Res. 141, 295–308.
Sumner A. 1972 A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 75, 304–306.
Tsipouri V., Schueler M. G., Hu S., NISC Comparative Sequencing Program, Dutra A., Pak E. et al. 2008 Comparative sequence analyses reveal sites of ancestral chromosomal fusions in the Indian muntjac genome. Genome Biol. 9, R155.
Wiens J. J., Kuczynski C. A., Hua X. and Moen D. S. 2010 An expanded phylogeny of treefrogs (Hylidae) based on nuclear and mitochondrial sequence data. Mol. Phylogenet. Evol. 55, 871–882.
Woznicki P., Sanchez L., Martinez P., Pardo B. G. and Jankun M. 2000 A population analysis of the structure and variability of NOR in Salmo trutta by Ag, CMA 3 and ISH. Genetica 108, 113–118.
Acknowledgements
We would like to express our thanks to two anonymous reviewers for helpful comments that improved the manuscript; Michele Orane, Luiz Fernando Kraft, Rafael Balen and Gisele Perazzo for help in lab work; the Grassmann family for permision to collect on their property; and Instituto Chico Mendes de Conservação da Biodiversidade, Instituto Ambiental do Paraná for environmental licenses. Besides, we are very grateful to Dr Mark A. McPeek for revising the English style of the manuscript. This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico eTecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado de São Paulo.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Oliveira I. S., Noleto R. B., Oliveira A. K. C., Toledo L. F. and Cestari M. M. 2016 Comparative cytogenetic analysis of four species of Dendropsophus (Hylinae) from the Brazilian Atlantic forest. J. Genet. 95, xx–xx]
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
SOARES DE OLIVEIRA, I., BUENO NOLETO, R., KARLOKOSKI CUNHA DE OLIVEIRA, A. et al. Comparative cytogenetic analysis of four species of Dendropsophus (Hylinae) from the Brazilian Atlantic forest. J Genet 95, 349–355 (2016). https://doi.org/10.1007/s12041-016-0645-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-016-0645-y