Abstract
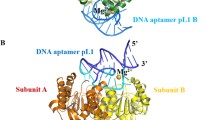
Bacillus halodurans (Bh) ribonuclease H (RNase H) belongs to the nucleotidyl-transferase (NT) superfamily and is a prototypical member of a large family of enzymes that use two-metal ion (Mg2+ or Mn2+) catalysis to cleave nucleic acids. Long timescale molecular dynamics simulations have been performed on the BhRNase H-DNA-RNA hybrid complex and the respective monomers to understand the recognition mechanism, conformational preorganization, active site dynamics and energetics involved in the complex formation. Several structural and energetic analyses were performed and significant structural changes are observed in enzyme and hybrid duplex during complex formation. Hybrid molecule binding to RNase H enzyme leads to conformational changes in the DNA strand. The ability of the DNA strand in the hybrid duplex to sample conformations corresponding to typical A- and B-type nucleic acids and the characteristic minor groove widthseem to be crucial for efficient binding. Sugar moieties in certain positions interacting with the protein structure undergo notable conformational transitions. The water coordination and arrangement around the metal ions in active site region are quite stable, suggesting their important role in enzymatic catalysis. Details of key interactions found at the interface of enzyme-nucleic acid complex that are responsible for its stability are discussed.

Molecular dynamics simulation results have shown that the DNA nucleotides of DNA-RNA hybrid that are interacting with ribonuclease H enzyme show interesting conformational changes upon binding.








Similar content being viewed by others
References
Nakamura H, Oda Y, Iwai S, Inoue H, Ohtsuka E, Kanaya S, Kimura S, Katsuda C, Katayanagi K and Morikawa K 1991 Proc. Natl. Acad. Sci. USA 88 11535
Nowotny M, Gaidamakov S A, Crouch R J and Yang W 2005 Cell 121 1005
Qiu J, Qian Y, Frank P, Wintersberger U and Shen B 1999 Mol. Cell. Biol. 19 8361
Crouch R J and Toulme J J 1998 In Ribonucleases H (INSERM Editions: Paris)
Tadokoro T and Kanaya S 2009 FEBS J. 276 1482
Worrall J A R and Luisi B F 2007 Curr. Opin. Struct. Biol. 17 128
Cerritelli S M and Crouch R J 2009 FEBS J. 276 1494
Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Ikehara M, Matsuzaki T and Morikawa K 1990 Nature 347 306
Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Nakamura H, Ikehara M, Matsuzaki T and Morikawa K 1992 J. Mol. Biol. 223 1029
Yang W, Hendrickson W A, Crouch R J and Satow Y 1990 Science 249 1398
Kanaya S, Kohara A, Miura Y, Sekiguchi A, Iwai S, Inoue H, Ohtsuka E and Ikehara M 1990 J. Biol. Chem. 265 4615
Haruki M, Noguchi E, Nakai C, Liu Y Y, Oobatake M, Itaya M and Kanaya S 1994 Eur. J. Bio. Chem. 220 623
Kanaya S and Crouch R J 1983 J. Biol. Chem. 258 1276
Oda Y, Iwai S, Ohtsuka E, Ishikawa M, Ikehara M and Nakamura H 1993 Nucleic Acid Res. 21 4690
Lai L, Yokota H, Hung L W, Kim R and Kim S H 2000 Struct. Fold. Des. 8 897
Ariyoshi M, Vassylyev D G, Iwasaki H, Nakamura H, Shinaweregawa H and Morikawa K 1994 Cell 78 1063
Ceschini S, Keeley A, McAlister M S, Oram M, Phelan J, Pearl L H, Tsaneva I R and Barrett T E 2001 EMBO J. 20 6601
Rice P A and Baker T A 2001 Nat. Struct. Biol. 8 302
Yang W and Steitz T A 1995 Structure 3 131
Gyi J I, Lane A N, Conn G L and Brown T 1998 Biochemistry 37 73
Suresh G and Priyakumar U D 2014 Phys. Chem. Chem. Phys. 16 18148
Nowotny M, Gaidamakov S A, Ghirlando R, Cerritelli S M, Crouch R J and Yang W 2007 Mol. Cell 28 264
Steitz T A and Steitz J A 1993 Proc. Natl. Acad. Sci. USA 90 6498
De Vivo M, Dal Peraro M and Klein M L 2008 J. Am. Chem. Soc. 130 10955
Krakowiak A, Owczarek A, Koziolkiewicz M and Stec W J 2002 Chembiochem 3 1242
Cassano A G, Anderson V E and Harris M E 2004 Biopolymers 73 110
Haruki M, Noguchi E, Kanaya S and Crouch R J 1997 J. Biol. Chem. 272 22015
Cerritelli S M, Frolova E G, Feng C, Grinberg A, Love P E and Crouch R J 2003 Mol. Cell 11 807
Luisi B F, Xu W X, Otwinowski Z, Freedman L P, Yamamoto K R and Sigler P B 1991 Nature 352 497
Nelson D L and Cox M M 2005 In Lehninger’s Principles of Biochemistry 4th ed. (New York: W H Freeman)
Draper D E 1993 Proc. Natl. Acad. Sci. USA 90 7429
Duan Y, Wilkosz P and Rosenberg J M 1996 J. Mol. Biol. 264 546
Arndt J W, Gong W, Zhong X, Showalter A K, Liu J, Dunlap C A, Lin Z, Paxson C, Tsai M D and Chan M K 2001 Biochemistry 40 5368
Tishchenko S, Nikonova E, Nikulin A, Nevskaya N, Volchkov S, Piendl W, Garber M and Nikonov S 2006 Acta Cryst. D 62 1545
Yang X, Gérczei T, Glover L and Correll C C 2001 Nature Struct. Biol. 8 968
Sen S and Nilsson L 1999 Biophys. J. 77 1782
Reyes C M, Nifosı R, Frankel A D and Kollman P A 2001 Biophys. J. 80 2833
Chen L, Zhang J, Yu L, Zheng Q C, Chu W T, Xue Q, Zhang H X and Sun C C 2012 J. Phys. Chem. B 116 12415
Driessen R P, Meng H, Suresh G, Shahapure R, Lanzani G, Priyakumar U D, White M F, Schiessel H, van Noort J and Dame R T 2013 Nucleic Acids Res. 41 196
Furini S, Barbini P and Domene C 2013 Nucleic Acids Res. 41 3963
Priyakumar U D, Harika G and Suresh G 2010 J. Phys. Chem. B 114 16548
Rosta E, Nowotny M, Yang W and Hummer G 2011 J. Am. Chem. Soc. 133 8934
Maláč K and Barvík I 2013 J. Mol. Graph. Model. 44 81
Brooks B R, Brooks C L, MacKerell A D, Nilsson L, Petrella R J, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner A R, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor R W, Post C B, Pu J Z, Schaefer M, Tidor B, Venable R M, Woodcock H L, Wu X, Yang W, York D M and Karplus M 2009 J. Comput. Chem. 30 1545
Priyakumar U D, Ramakrishna S, Nagarjuna K R and Reddy S K 2010 J. Phys. Chem. B 114 1707
Suresh G and Priyakumar U D 2014 J. Phys. Chem. B 118 5853
Jorgensen W L, Chandrasekhar J, Madura J D, Impey R W and Klein M L 1983 J. Chem. Phys. 79 926
Ryckaert J P, Ciccotti G and Berendsen H J C 1977 J. Comput. Phys. 23 327
Field M J and Karplus M 1992 In CRYSTAL: Program for Crystal Calculations in CHARMM (Harvard University: Cambridge, MA)
Darden T, Perera L, Li L P and Pedersen L 1999 Structure 7 R55
Essmann U, Perera L, Berkowitz M L, Darden T A, Lee H and Pedersen L G 1995 J. Chem. Phys. 103 8577
Steinbach P J and Brooks B R 1994 J. Comput. Chem. 15 667
Phillips J C, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel R D, Kale L and Schulten K 2005 J. Comput. Chem. 26 1781
Foloppe N and MacKerell A D 2000 J. Comput. Chem. 21 86
MacKerell A D and Banavali N K 2000 J. Comput. Chem. 21 105
MacKerell A D, Bashford D, Bellott D R L, Evanseck J D, Field M J, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau F T K, Mattos C, Michnick S, Ngo T, Nguyen D T, Prodhom B, Reiher W E, Roux B, Schlenkrich M, Smith J C, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D and Karplus M 1998 J. Phys. Chem. B 102 3586
MacKerell A D, Feig M and Brooks C L 2004 J. Comput. Chem. 25 1400
Suresh G and Priyakumar U D 2013 J. Phys. Chem. B 117 5556
Feller S E, Zhang Y, Pastor R W and Brooks R W 1995 J. Chem. Phys. 103 4613
Hoover W G 1985 Phy. Rev. A 31 1695
Priyakumar U D and MacKerell A D 2010 J. Mol. Biol. 396 1422
Humphrey W, Dalke A and Schulten K 1996 J. Molec. Graphics 14 33
Lavery R, Moakher M, Maddocks J H, Petkeviciute D and Zakrzewska K 2009 Nucleic Acids Res. 37 5917
Stafford K A and Palmer A G 2014 F1000 Research 3 67
Rychlik M P, Chon H, Cerritelli S M, Klimek P, Crouch R J and Nowotny M 2010 Molecular Cell 40 658
Babu C S, Dudev T and Lim C 2013 J. Am. Chem. Soc. 135 6541
Acknowledgements
We thank AICTE, and Department of Atomic Energy-BRNS (37(2)/14/05/2015/BRNS/20046) for financial assistance. GS thanks Council of Scientific and Industrial Research (CSIR), India for senior research fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information
This includes methods to calculate hydration numbers and hydrogen bonds (Supplementary methods), RMSD and radius of gyration (figure S1; table S1), Ramachandran plot (figure S2), hydrogen bond distances (figure S3), helical parameters (figures S4–S5), backbone angles (figure S6), buried surface area (figure S7), hydrogen bond distances (figure S8), pseudorotation angles (table S2), stacking interactions (table S3), hydration numbers and sasa (table S4), and statistics of hydrogen bonds (tables S5, S6, S7). Supplementary Information is available at www.ias.ac.in/chemsci.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
SURESH, G., PRIYAKUMAR, U.D. Atomistic details of the molecular recognition of DNA-RNA hybrid duplex by ribonuclease H enzyme. J Chem Sci 127, 1701–1713 (2015). https://doi.org/10.1007/s12039-015-0942-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12039-015-0942-7