Abstract
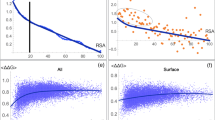
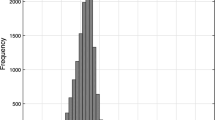
Understanding the features of the protein conformational space represents a key component to characterize protein structural evolution at the molecular level. This problem is approached in a twofold manner; simple lattice models are used to represent protein structures with the ability of a protein sequence to fold into the lowest energy native conformation, quantified as the foldability, which measures the fitness of the sequence. Alternatively, a self-consistent mean-field based theory is developed to evaluate the protein neutrality through random single-point and multiple-point mutations by calculating the pair-wise probability profile of the amino acid residues in a library of sequences, consistent with a particular foldability criterion. The theory predicts the change in sequence plasticity with the foldability criterion and also correlates the effect of hydrophobic residues on the variation of the free energy surface of the protein as a function of the number of cumulative mutations. The results obtained from the theory are compared with the exact enumeration results of 3 × 3 × 3 lattice protein and also with some small real proteins chosen from the protein databank. An excellent match of the results obtained from theory and exact enumeration with those of real proteins validates the range of applicability of the theory. The theory may provide a new perspective in de novo protein design, in-vivo/in-vitro protein evolution and site-directed mutagenesis experiments.
Similar content being viewed by others
References
Schuster P, Fontana W, Stadler P F and Hofacker I L 1994 Proc. Roy. Soc. London B255 279
Govindarajan S and Goldstein R A 1997 Biopolymers 42 427
Bornberg-Bauer E and Chan H S 1999 Proc. Natl. Acad. Sci. USA 96 10689
vanNimwegen E, Crutchfiels J P and Huynen M 1999 Proc. Natl. Acad. Sci. USA 96 9716
Bastolla U, Porto M, Roman H E and Vendruscolo M 2002 Phys. Rev. Lett. 89 208101
Thomas J, Martin O C and Wagner A 2008 BMC Bioinformatics 9 464
Reidys C, Stadler P F and Schuster P 1997 Bull. Math. Biol. 59 339
Schuster P 1995 J. Biotechnol. 41 239
Sali A, Shaknovich E and Karplus M 1994 J. Mol. Biol. 235 1614
Shakhnovich E I and Gutin A M 1990 J. Chem. Phys. 93 5967
Bryngelson J D and Wolynes P G 1987 Proc. Natl. Acad. Sci. USA 84 7524
Martinez M A, Pezo V, Marliere P and Wain-Hobson S 1996 The EMBO J. 15 1203
Shortle D and Lin B 1985 Genetics 110 539
Pakula A A, Young V B and Sauer R T 1986 Proc. Natl. Acad. Sci. USA 83 8829
Guo H H, Choe J and Loeb L A 2004 Proc. Natl. Acad. Sci. USA 101 9205
Bloom J D, Labthavikul S T, Otey C R and Arnold F H 2006 Proc. Natl. Acad. Sci. USA 103 5869
Serrano L, Day A G and Fersht A R 1993 J. Mol. Biol. 233 305
King J L and Jukes T H 1969 Science 164 788
Kimura M 1983 The neutral theory of molecular evolution (Cambridge University Press)
Bloom J D, Silberg J J, Wilke C O, Drummond D A, Adami C and Arnold F H 2005 Proc. Natl. Acad. Sci. USA 606 102
Go N 1983 Annu. Rev. Biophys. Bioeng. 12 183
Li H, Helling R, Tang C and Wingreen N 1996 Science 273 666
Russ W P and Ranganathan R 2002 Curr. Opin. Struct. Biol. 12 447
Go N and Taketomi H 1978 Proc. Natl. Acad. Sci. USA 75 559
Chan H S and Dill K A 1991 Annu. Rev. Biophys. Biophys. Chem. 20 447
Shaknovich E I 1994 Phys. Rev. Lett. 72 3907
Socci N D and Onuchic J N 1995 J. Chem. Phys. 103 4732
Abkevich V I, Gutin A M and Shakhnovich E I 1995 Protein. Sci. 4 1167
Hao M-H and Scheraga H A 1996 Proc. Natl. Acad. Sci. USA 93 4984
Zou J and Saven J G 2000 J. Mol. Biol. 296 281
Biswas P, Zou J and Saven J G 2005 J. Chem. Phys. 123 154908
Miyazawa S and Jernigan R L 1985 Macromolecules 218 534
Sippl M J 1990 J. Mol. Biol. 213 859
Goldstein R, Luthey-Schulten Z Z and Wolynes P G 1992 Proc. Natl. Acad. Sci. USA 89 9029
Deutsch J M and Kurosky T 1996 Phys. Rev. Lett. 76 323
Saven J G 2003 J. Chem. Phys. 118 6133
Morita T and Tanaka T 1966 Phys. Rev. 145 288
Pathria R K 1972 Statistical mechanics (Pergamon Press)
Shaknovich E I and Gutin A 1990 J. Chem. Phys. 93 5967
Go N 1975 Int. J. Pept. Protein Res. 7 313
Dill K A, Bromberg S, Yue K S, Fiebig K M, Yee D P, Thomas P D and Chan H S 1995 Protein Sci. 4 561
Saven J G 2001 Chem. Rev. (Washington DC) 101 3113
Bhattacherjee A and Biswas P 2009 J. Phys. Chem. B (ASAP), DOI 10.1021/jp810515s
Dosztanyi Z, Csizmok V, Tompa P and Simon I 2005 J. Mol. Biol. 347 827
Sharp K A, Nicholls A, Friedmann R and Honig B 1991 Biochemistry 30 9686
Levitt M 1976 J. Mol. Biol. 104 59
Zhou H and Zhou Y 2002 Proteins: Struct. Funct. Genet. 49 483
Fauchere J-L and Pliska V 1983 Eur. J. Med. Chem. (Chim. Ther.) 18 369
Taverna D M and Goldstein R A 2002 J. Mol. Biol. 315 479
Oliveira L C, Silva R T H, Leite V B P and Chahine J 2006 J. Chem. Phys. 125 084904
Wilke C O, Bloom J D, Drummond D A and Raval A 2005 Biophys. J. 89 3714
Sano K-I, Maeda K, Taniguchi H and Maeda Y 2000 Eur. J. Biochem. 267 4870
Miao J, Klein-Seetharaman J and Meirovitch H 2004 J. Mol. Biol. 344 797
Chan K A and Dill H S 1994 J. Chem. Phys. 100 9238
Hamming R W 1950 Bell. Syst. Tech. J. 29 147
Hamming R W 1986 Coding and information theory (Englewood Cliffs: Prentice Hall) 2nd edn
Hecht M H, Hehir K M, Nelson H C M, Sturtevant J M and Sauer R T 1985 J. Cell. Biochem. 29 217
Hecht M H, Sturtevant J M and Sauer R T 1986 Proteins. Struct. Funct. Genet. 1 43
Irback A and Troein C 2002 J. Biol. Phys. 28 1
Chan H S and Dill K A K 1996 Proteins: Struct. Funct. Genet. 24 335
Author information
Authors and Affiliations
Corresponding author
Additional information
Dedicated to the memory of the late Professor S K Rangarajan
Rights and permissions
About this article
Cite this article
Bhattacherjee, A., Biswas, P. Statistical theory of neutral protein evolution by random site mutations. J Chem Sci 121, 887–896 (2009). https://doi.org/10.1007/s12039-009-0105-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12039-009-0105-9