Abstract
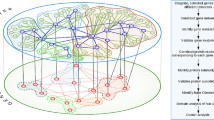
The role played by the topological structure of biological networks in their dynamics and function is receiving increasing attention over the last decade as large-throughput experiments have provided large volumes of highly resolved data on the interactions between the components of such networks. This has provided new perspectives on systems diseases: for example, there has been a gradual shift in cancer research away from the study of individual molecules and of single gene mutations to the emerging consensus that it is a complex disease involving large-scale disruptions in the intracellular signaling network. One of the drawbacks of a systems- or network-based approach is the large number of cellular agents whose interactions need to be investigated. We tried to solve this problem by taking a mesoscopic view of the cancer diseases–genes network, whose modular organization we studied after projecting it onto two networks, one comprising only disease types and the other consisting of only genes related to one or more categories of cancer. Using community partitioning, we identified several modules in these networks. Projecting cancer gene clusters onto an abstract ‘modular space’ allows us to infer the relations between different tumor types. By classifying the functional role of particular genes in terms of their inter- and intra-modular connectivity, we identified a number of genes that play the key role of ‘connector hubs’ in the network. Using data from the human protein–protein interaction network we showed that genes that are ‘connector hubs’ or ‘global hubs’ are, in fact, much more likely to be related to cancer than other genes. More important from a therapeutic point of view, we showed that the connector hubs in the cancer gene network are involved in a significantly larger number of human signaling pathways associated with cancer than other types of cancer genes. Furthermore, the types of cancer linked to connector hub genes have significantly reduced survival rates compared with other types of cancer, thereby enhancing their importance in the search for potential therapeutic targets.








Similar content being viewed by others
References
Clauset A, Newman ME and Moore C 2004 Finding community structure in very large networks. Phys. Rev. E 70 066111
Cloutier M and Wang E 2011 Dynamic modeling and analysis of cancer cellular network motifs. Integr. Biol. 3 724–732
Fortunato S 2010 Community detection in graphs. Phys. Rep. 486 75–174
Futreal AP, Coin L, Marshall M, et al. 2006 A census of human cancer genes. Nat. Rev. Cancer 4 177–183
Goh KI, Cusick ME, Valle D, et al. 2007 The human disease network. Proc. Natl. Acad. Sci. USA 104 8685–8690
Gong X, Wu R, Zhang Y, et al. 2010 Extracting consistent knowledge from highly inconsistent cancer gene data sources. BMC Bioinform. 11 76
Good BH, De Montjoye YA and Clauset A 2010 Performance of modularity maximization in practical contexts. Phys. Rev. E 81 046106
Guimera R and Amaral LAN 2005 Cartography of complex networks: modules and universal roles. J. Stat. Mech.. https://doi.org/10.1088/1742-5468/2005/02/P02001
Guimera R, Pardo MS and Amaral LAN 2007 Classes of complex networks defined by role-to-role connectivity profiles. Nat. Phys. 3 63–69
Hamosh A, Scott AF, Amberger JS, Bocchini CA and McKusick VA 2005 Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33 D514–D517
Hartwell LH, Hopfield JJ, Leibler S and Murray AW 1999 From molecular to modular cell biology. Nature 402 C47–C52
Hornberg JJ, Bruggeman FJ, Westerhoff HV and Lankelma J 2006 Cancer: a systems biology disease. Biosystems 83 81–90
Kreeger PK and Lauffenburger DA 2010 Cancer systems biology: a network modeling perspective. Carcinogenesis 31 2–8
Lancichinetti A and Fortunato S 2009 Community detection algorithms: a comparative analysis. Phys. Rev. E 80 056117
Lancichinetti A and Fortunato S 2012 Consensus clustering in complex networks. Sci. Rep. 2 336
Lee JA and Verleysen M 2007 Nonlinear dimensionality reduction (New York: Springer)
Mariotto AB, Noone AM, Howlader N, et al. 2014 Cancer survival: an overview of measures, uses, and interpretation. J. Nat. Cancer Inst. Monogr. 2014 145–186
Milo R, Shen-Orr S, Itzkovitz S, et al. 2002 Network motifs: simple building blocks of complex networks. Science 298 824–827
Mukherjee S 2010 The emperor of all maladies: A biography of cancer (New York: Scribner)
Newman MEJ 2004 Detecting community structure in networks. Eur. Phys. J. B 38 321–330
Newman MEJ 2006 Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 103 8577–8582
Newman MEJ and Girvan M 2004 Finding and evaluating community structure in networks. Phys. Rev. E 69 026113
Porter MA, Onnela JP and Mucha PJ 2009 Communities in Networks. Notices AMS 56 1082–1097
Prasad TSK, Goel R, Kandasamy K, et al. 2009 Human Protein Reference Database - 2009 update. Nucleic Acids Res. 37 D767–D772
Rosvall M and Bergstrom CT 2008 Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. USA 105 1118–1123
Rosvall M and Bergstrom CT 2010 Mapping change in large networks. PLoS One 5 e8694
Schaefer CF, Anthony K, Krupa S, et al. 2009 PID: the pathway interaction database. Nucleic Acids Res. 37 D674–D679
Schaub MT, Delvenne JC, Rosvall M and Lambiotte R 2017 The many facets of community detection in complex networks. Appl. Netw. Sci. 2 4
WHO 2021 Cancer fact sheet https://www.who.int/en/news-room/fact-sheets/detail/cancer
Acknowledgements
The work was partly supported by the IMSc Complex Systems Project (XII Plan) funded by the Department of Atomic Energy, Government of India. We would like to thank Soumya Easwaran and Anand Pathak for their assistance in preparing figures, and Indrani Bose, Shaon Chakrabarti, Arjun Krishnan, Ramakrishna Ramaswamy, M S Santhanam and Somdatta Sinha for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Mohit Kumar Jolly.
Corresponding editor: Mohit Kumar Jolly
This article is part of the Topical Collection: Emergent dynamics of biological networks.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Flowchart indicating schematically the methods employed for the analysis reported in the paper (TIF 3275 kb)
Supplementary Figure 2
Visual representation of the comparison between the modular decomposition of the TT-GWN obtained using Infomap method (top, 25 modules) with that obtained using spectral partitioning (bottom, 13 modules). The modules are represented as horizontal line segments, connected by bands which are colored according to the module (obtained using the Infomap method) from which they originate (TIF 641 kb)
Supplementary Figure 3
The composition of the modules of TT-GWN in terms of different cellular components (TIF 1650 kb)
Supplementary Figure 4
: The composition of the modules of TT-GWN in terms of different biological processes (TIF 1494)
Supplementary Figure 5
Overlap in terms of gene memberships of different cancer categories (shown along the abscissae) and (shown along the ordinate) different (a) human signaling pathways that are related to cancer [obtained from the National Cancer Institute (NCI) database], (b) cellular components and (c) biological processes, obtained from Gene Ontology (GO). Overlap (i,j) is measured by the fraction of genes in a particular cancer category j for which the corresponding proteins that they express occur in the (a) signaling pathway i, or (b) cellular component i, or (c) biological process i. The extent of overlap is represented using the colorbar shown at right (TIF 317 kb)
Rights and permissions
About this article
Cite this article
Jesan, T., Sinha, S. Modular organization of gene–tumor association network allows identification of key molecular players in cancer. J Biosci 47, 60 (2022). https://doi.org/10.1007/s12038-022-00292-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12038-022-00292-5