Abstract
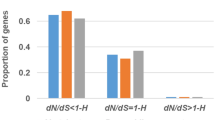
Matrix metalloproteinases-9 (MMP-9) is an important cancer-associated, zinc-dependent endopeptidase. To investigate the natural selection hypothesis of MMP-9, the orthologous sequences from 12 vertebrates were compared and a molecular evolution analysis was performed. Results suggest that amino acid residues present in the middle region of the protein are more selectively constrained, whereas amino acid residues in the C-terminal region of the MMP-9 protein including exon 13 showed lowest conservation level in non-primate species, suggesting that it is an exon with fast evolving rate compared to the others analyzed. InterProScan analysis shows that exon 13 was located in hemopexin (PEX) domain of MMP-9. Positive selection was detected in PEX domain of MMP-9 protein between human and other species, which indicates that selective pressure may play a role in shaping the function of MMP-9 in the course of evolution.




Similar content being viewed by others
References
Bjorklund M, Heikkila P and Koivunen E 2004 Peptide inhibition of catalytic and noncatalytic activities of matrix metalloproteinase-9 blocks tumor cell migration and invasion. J. Biol. Chem. 279 29589–29597
Burg-Roderfeld M, Roderfeld M, Wagner S, Henkel C, Grotzinger J and Roeb E 2007 MMP-9-hemopexin domain hampers adhesion and migration of colorectal cancer cells. Int. J. Oncol. 30 985–992
Cha H, Kopetzki E, Huber R, Lanzendorfer M and Brandstetter H 2002 Structural basis of the adaptive molecular recognition by MMP9. J. Mol. Biol. 320 1065–1079
Cruciani V and Mikalsen SO 2007 Evolutionary selection pressure and family relationships among connexin genes. Biol. Chem. 388 253–264
Massova I, Kotra LP, Fridman R and Mobashery S 1998 Matrix metalloproteinases: structures, evolution, and diversification. FASEB J. 12 1075–1095
Morgunova E, Tuuttila A, Bergmann U and Tryggvason K 2002 Structural insight into the complex formation of latent matrix metalloproteinase 2 with tissue inhibitor of metalloproteinase 2. Proc. Nat. Acad. Sci. USA 99 7414–7419
Mulder N and Apweiler R 2007 InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol. Biol. 396 59–70
Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA and O'Brien SJ 2001 Molecular phylogenetics and the origins of placental mammals. Nature 409 614–618
Nagase H, Visse R and Murphy G 2006 Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 69 562–573
Ohta T 1992 The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23 263–286
Piccard H, Van den Steen PE and Opdenakker G 2007 Hemopexin domains as multifunctional liganding modules in matrix metalloproteinases and other proteins. J. Leukoc. Biol. 81 870–892
Rawlings ND, Barrett AJ and Bateman A 2010 MEROPS: the peptidase database. Nucleic Acids Res. 38 D227–233
Tamura K, Peterson D, Peterson N, Stecher G, Nei M and Kumar S 2011 MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739
Tolosano E and Altruda F 2002 Hemopexin: structure, function, and regulation. DNA Cell Biol. 21 297–306
Vandooren J, Van den Steen PE and Opdenakker G 2013 Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): the next decade. Crit. Rev. Biochem. Mol. Biol. 48 222–272
Yang Z 2007 PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 1586–1591
Acknowledgements
This work was sponsored by National Natural Science Foundation of China (grant no. 31301486) and Natural Science Foundation of Shanghai (grant no. 13ZR1439600).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Corresponding editor: Stuart A. Newman
[Liu Y, Zhao Y, Lu C, Fu M, Dou T and Tan X 2015 Signatures of positive selection at hemopexin (PEX) domain of matrix metalloproteinase-9 (MMP-9) gene. J. Biosci.] DOI 10.1007/s12038-015-9577-6
Rights and permissions
About this article
Cite this article
Liu, Y., Zhao, Y., Lu, C. et al. Signatures of positive selection at hemopexin (PEX) domain of matrix metalloproteinase-9 (MMP-9) gene. J Biosci 40, 885–890 (2015). https://doi.org/10.1007/s12038-015-9577-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12038-015-9577-6