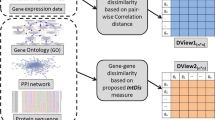
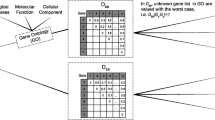
Abstract
A challenge in bioinformatics is to analyse volumes of gene expression data generated through microarray experiments and obtain useful information. Consequently, most microarray studies demand complex data analysis to infer biologically meaningful information from such high-throughput data. Selection of informative genes is an important data analysis step to identify a set of genes which can further help in finding the biological information embedded in microarray data, and thus assists in diagnosis, prognosis and treatment of the disease. In this article we present an unsupervised feature selection technique which attempts to address the goal of explorative data analysis, unfolding the multi-faceted nature of data. It focuses on extracting multiple clustering views considering the diversity of each view from high-dimensional data. We evaluated our technique on benchmark data sets and the experimental results indicates the potential and effectiveness of the proposed model in comparison to the traditional single view clustering models, as well as other existing methods used in the literature for the studied datasets.




Similar content being viewed by others
References
Berriz GF, Beaver JE, Cenik C, Tasan M and Roth FP 2009 Next generation software for functional trend analysis. Bioinformatics 25 3043–3044
Bickel S and Scheffer T 2004 Multi-view clustering; in Proceedings of the Fourth IEEE International Conference on Data Mining, ICDM '04, IEEE Computer Society pp 19–26
Boutsidis C, et al. 2008 Unsupervised feature selection for principal components analysis; in Proc. of the 14th ACM SIGKDD Int. Conf. on Knowledge discovery and data mining pp 61–69
Bruno E and Marchand-Maillet S 2009 Multiview clustering: a late fusion approach using latent models; in SIGIR (ACM) pp 736–737
Chaudhuri K, Kakade SM, Livescu K, and Sridharan K 2009 Multi-view clustering via canonical correlation analysis; in Proceedings of the 26th Annual International Conference on Machine Learning, ICML '09 pp 129–136
Chen X, Xu X, Huang J and Ye Y 2013 TW-(k)-means: automated two-level variable weighting clustering algorithm for multiview data. IEEE Trans. Knowl. Data Eng. 25 932–944
Cho JH, Gelinas R, Wang K, Etheridge A, Piper MG, Batte K, Dakhlallah D, Price J, et al. 2011 Systems biology of interstitial lung diseases: integration of mrna and microrna expression changes. BMC Med. Genet. 4 8
Chuang HY, Rassenti L, Salcedo M, Licon K, Kohlmann A, Haferlach T, Foà R, Ideker T, et al. 2012 Subnetwork-based analysis of chronic lymphocytic leukemia identifies pathways that associate with disease progression. Blood 120 2639–2649
Codony C, Crespo M, Abrisqueta P, Montserrat E and Bosch F 2009 Gene expression profiling in chronic lymphocytic leukaemia. Best Pract. Res. Clin. Haematol. 22 211–222
Cottin V 2013 Interstitial lung disease. Eur. Respir. Rev. 22 26–32
Cui Y, Fern, XZ, Dy and JG 2007 Non-redundant multi-view clustering via orthogonalization. Proc. 7th IEEE International Conference on Data Mining (ICDM'07) pp 133–142
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA, et al. 2003 David: database for annotation, visualization, and integrated discovery. Genome Biol. 4 P3
Di W and Crawford MM 2012 View generation for multiview maximum disagreement based active learning for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 50 1942–1954
Ding CH 2003 Unsupervised feature selection via two-way ordering in gene expression analysis. Bioinformatics 19 1259–1266
Dudoit S, Yang YH, Callow MJ and Speed TP 2002 Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat. Sin. 12 111–140
Fält S, Merup M, Gahrton G, Lambert B and Wennborg A 2005 Identification of progression markers in b-cll by gene expression profiling. Exp. Hematol. 33 883–893
Fang G, Kuang R, Pandey G, Steinbach M, Myers CL and Kumar V 2010 Subspace differential coexpression analysis: problem definition and a general approach; in Pacific Symp Biocomput 15 145–56
Ghosh A, Dhara BC and De RK 2014 Selection of genes mediating certain cancers, using a neuro fuzzy approach. Neurocomputing 133 122–140
Gupta R, Rao N and Kumar V 2011 Discovery of error-tolerant biclusters from noisy gene expression data. BMC Bioinforma. 12 S1
Hong Y, Kwong S, Chang Y and Ren Q 2008 Consensus unsupervised feature ranking from multiple views. Pattern Recogn. Lett. 29 595–602
Huang DW, Sherman BT and Lempicki RA 2008 Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 4 44–57
Huang DW, Sherman BT and Lempicki RA 2009 Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 1–13
Jaeger J, et al. 2003 Improved gene selection for classification of microarrays; in Pacific Symp. on Biocomputing pp 53–64
Ji J, Zhang A, Liu C, Quan X and Liu Z 2014 Survey: functional module detection from protein-protein interaction networks. IEEE Trans. Knowl. Data Eng. 26 261–277
Kim YM, Amini MR, Goutte C and Gallinari P 2010 Multi-view clustering of multilingual documents; in SIGIR (ACM) pp 821–822
Kohane IS, Butte AJ and Kho A 2002 Microarrays for an integrative genomics. MIT press
Li G, et al. 2008 A novel unsupervised feature selection method for bioinformatics data sets through feature clustering; in 2008 I.E. Int. Conf. on Granular Comput pp 41–47
Mitra P and Swarnkar T 2012 Graph based unsupervised feature selection for microarray data; in Proceedings of the 2012 I.E. International Conference on Bioinformatics and Biomedicine Workshops (BIBMW), IEEE Computer Society pp 750–751
Mitra P, Murthy CA and Pal SK 2002 Unsupervised feature selection using feature similarity. IEEE Trans. Pattern Anal. Mach. Intell. 24 301–312
Muller E, Gunnemann S, Farber I and Seidl T 2012 Discovering multiple clustering solutions: Grouping objects in different views of the data; in IEEE 28th International Conference on Data Engineering, IEEE pp 1207–1210
Pirim H, Ekiolu B, Perkins AD and Yüceer C 2012 Clustering of high throughput gene expression data. Comput. Oper. Res. 39 3046–3061
Safran M, Dalah I, Alexander J, Rosen N, Stein TI, Shmoish M, Nativ N, Bahir I, et al. 2010 GeneCards version 3: the human gene integrator. doi:10.1093/database/baq020
Sharma A, Imoto S and Miyano S 2012a A filter based feature selection algorithm using null space of covariance matrix for dna microarray gene expression data. Curr. Bioinforma 7 289–294
Sharma A, Imoto S and Miyano S 2012b A top-r feature selection algorithm for microarray gene expression data. IEEE/ACM Trans. Comput. Biol. Bioinformatics 9 754–764
Sun S 2013 A survey of multi-view machine learning. Neural Comput. Applic. 23 2031–2038
Swarnkar T, Simões SN, Martins-Jr DC, Anura A, Brentani H, Hashimoto RF and Mitra P 2014 Multiview clustering on ppi network for gene selection and enrichment from microarray data. In: IEEE International Conference on BioInformatics and BioEngineering
Tibshirani R and Witten D 2007 A comparison of fold-change and the t-statistic for microarray data analysis. Technical report, Stanford, CA: Stanford University
Varshavsky R, Gottlieb A, Linial M and Horn D 2006 Novel unsupervised feature filtering of biological data. Bioinformatics 22 e507–e513
Xiao Y, Hsiao TH, Suresh U, Chen HIH, Wu X, Wolf SE and Chen Y 2014 A novel significance score for gene selection and ranking. Bioinformatics 30 801–807
Xu C, Tao D and Xu C 2013 A survey on multi-view learning. arXiv preprint arXiv:1304.5634
Yang P, Hwa Yang Y, Zhou BB and Zomaya YA 2010 A review of ensemble methods in bioinformatics. Curr. Bioinforma. 5 296–308
Yu L and Liu H 2004 Efficient feature selection via analysis of relevance and redundancy. J. Mach. Learn. Res. 5 1205–1224
Yu S, Tranchevent LC, De Moor B and Moreau Y 2010 Gene prioritization and clustering by multi-view text mining. BMC Bioinf. 11 28
Author information
Authors and Affiliations
Corresponding author
Additional information
[Swarnkar T and Mitra P 2015 Graph-based unsupervised feature selection and multiview clustering for microarray data. J. Biosci.] DOI 10.1007/s12038-015-9559-8
Rights and permissions
About this article
Cite this article
Swarnkar, T., Mitra, P. Graph-based unsupervised feature selection and multiview clustering for microarray data. J Biosci 40, 755–767 (2015). https://doi.org/10.1007/s12038-015-9559-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12038-015-9559-8