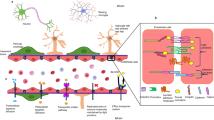
Abstract
Major depressive disorder (MDD) is a severe mental illness characterized by a lack of objective biomarkers. Mounting evidence suggests there are extensive transcriptional molecular changes in the prefrontal cortex (PFC) of individuals with MDD. However, it remains unclear whether there are specific genes that are consistently altered and possess diagnostic power. In this study, we conducted a systematic search of PFC datasets of MDD patients from the Gene Expression Omnibus database. We calculated the differential expression of genes (DEGs) and identified robust DEGs using the RRA and MetaDE methods. Furthermore, we validated the consistently altered genes and assessed their diagnostic power through enzyme-linked immunosorbent assay experiments in our clinical blood cohort. Additionally, we evaluated the diagnostic power of hub DEGs in independent public blood datasets. We obtained eight PFC datasets, comprising 158 MDD patients and 263 healthy controls, and identified a total of 1468 unique DEGs. Through integrated analysis, we identified 290 robustly altered DEGs. Among these, seven hub DEGs (SLC1A3, PON2, AQP1, EFEMP1, GJA1, CENPD, HSD11B1) were significantly down-regulated at the protein level in our clinical blood cohort. Moreover, these hub DEGs exhibited a negative correlation with the Hamilton Depression Scale score (P < 0.05). Furthermore, these hub DEGs formed a panel with promising diagnostic power in three independent public blood datasets (average AUCs of 0.85) and our clinical blood cohort (AUC of 0.92). The biomarker panel composed of these genes demonstrated promising diagnostic efficacy for MDD and serves as a useful tool for its diagnosis.




Similar content being viewed by others
Data Availability
The data in this study was the public available datasets, which can be found in Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database.
References
Marx W, Penninx B, Solmi M, Furukawa TA, Firth J, Carvalho AF, Berk M (2023) Major depressive disorder. Nat Rev Dis Primers 9(1):44. https://doi.org/10.1038/s41572-023-00454-1
Dahl A, Thompson M, An U, Krebs M, Appadurai V, Border R, Bacanu SA, Werge T et al (2023) Phenotype integration improves power and preserves specificity in biobank-based genetic studies of major depressive disorder. Nat Genet 55(12):2082–2093. https://doi.org/10.1038/s41588-023-01559-9
Friedrich MJ (2017) Depression is the leading cause of disability around the world. JAMA 317(15):1517. https://doi.org/10.1001/jama.2017.3826
Trivedi MH, Rush AJ, Wisniewski SR, Nierenberg AA, Warden D, Ritz L, Norquist G, Howland RH, Team SDS et al (2006) Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry 163(1):28–40. https://doi.org/10.1176/appi.ajp.163.1.28
Wang Y, Zhou J, Ye J, Sun Z, He Y, Zhao Y, Ren S, Zhang G et al (2023) Multi-omics reveal microbial determinants impacting the treatment outcome of antidepressants in major depressive disorder. Microbiome 11(1):195. https://doi.org/10.1186/s40168-023-01635-6
Ho CSH, Wang J, Tay GWN, Ho R, Husain SF, Chiang SK, Lin H, Cheng X et al (2024) Interpretable deep learning model for major depressive disorder assessment based on functional near-infrared spectroscopy. Asian J Psychiatr 92:103901. https://doi.org/10.1016/j.ajp.2023.103901
Chen Q, Liu W, Li H, Zhang H, Tian M (2011) Molecular imaging in patients with mood disorders: a review of PET findings. Eur J Nucl Med Mol Imaging 38(7):1367–1380. https://doi.org/10.1007/s00259-011-1779-z
Hidano A, Mita K (1966) Ichthyosis-like changes of the skin and internal diseases with special reference to complications of malignant tumors. Nihon Rinsho 24(2):250–254
Belleau EL, Treadway MT, Pizzagalli DA (2019) The impact of stress and major depressive disorder on hippocampal and medial prefrontal cortex morphology. Biol Psychiatry 85(6):443–453. https://doi.org/10.1016/j.biopsych.2018.09.031
Kang HJ, Voleti B, Hajszan T, Rajkowska G, Stockmeier CA, Licznerski P, Lepack A, Majik MS et al (2012) Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat Med 18(9):1413–1417. https://doi.org/10.1038/nm.2886
Nagy C, Maitra M, Tanti A, Suderman M, Theroux JF, Davoli MA, Perlman K, Yerko V et al (2020) Single-nucleus transcriptomics of the prefrontal cortex in major depressive disorder implicates oligodendrocyte precursor cells and excitatory neurons. Nat Neurosci 23(6):771–781. https://doi.org/10.1038/s41593-020-0621-y
Huang Y, Wu J, Zhang H, Li Y, Wen L, Tan X, Cheng K, Liu Y et al (2023) The gut microbiome modulates the transformation of microglial subtypes. Mol Psychiatry 28(4):1611–1621. https://doi.org/10.1038/s41380-023-02017-y
Wu J, Li Y, Huang Y, Liu L, Zhang H, Nagy C, Tan X, Cheng K et al (2023) Integrating spatial and single-nucleus transcriptomic data elucidates microglial-specific responses in female cynomolgus macaques with depressive-like behaviors. Nat Neurosci 26(8):1352–1364. https://doi.org/10.1038/s41593-023-01379-4
Scarpa JR, Fatma M, Loh YE, Traore SR, Stefan T, Chen TH, Nestler EJ, Labonte B (2020) Shared transcriptional signatures in major depressive disorder and mouse chronic stress models. Biol Psychiatry 88(2):159–168. https://doi.org/10.1016/j.biopsych.2019.12.029
Cipriani A, Zhou X, Del Giovane C, Hetrick SE, Qin B, Whittington C, Coghill D et al (2016) Comparative efficacy and tolerability of antidepressants for major depressive disorder in children and adolescents: a network meta-analysis. Lancet 388(10047):881–890. https://doi.org/10.1016/S0140-6736(16)30385-3
Liu L, Wang H, Zhang H, Chen X, Zhang Y, Wu J, Zhao L, Wang D et al (2022) Toward a deeper understanding of gut microbiome in depression: the promise of clinical applicability. Adv Sci (Weinh) 9(35):e2203707. https://doi.org/10.1002/advs.202203707
Pu J, Liu Y, Zhang H, Tian L, Gui S, Yu Y, Chen X, Chen Y et al (2021) An integrated meta-analysis of peripheral blood metabolites and biological functions in major depressive disorder. Mol Psychiatry 26(8):4265–4276. https://doi.org/10.1038/s41380-020-0645-4
Pu J, Liu Y, Gui S, Tian L, Yu Y, Song X, Zhong X, Chen X et al (2021) Metabolomic changes in animal models of depression: a systematic analysis. Mol Psychiatry 26(12):7328–7336. https://doi.org/10.1038/s41380-021-01269-w
Shi Y, Song R, Wang L, Qi Y, Zhang H, Zhu J, Zhang X, Tang X et al (2020) Identifying Plasma Biomarkers with high specificity for major depressive disorder: a multi-level proteomics study. J Affect Disord 277:620–630. https://doi.org/10.1016/j.jad.2020.08.078
Torres-Berrio A, Morgunova A, Giroux M, Cuesta S, Nestler EJ, Flores C (2021) miR-218 in adolescence predicts and mediates vulnerability to stress. Biol Psychiatry 89(9):911–919. https://doi.org/10.1016/j.biopsych.2020.10.015
Torres-Berrio A, Nouel D, Cuesta S, Parise EM, Restrepo-Lozano JM, Larochelle P, Nestler EJ, Flores C (2020) MiR-218: a molecular switch and potential biomarker of susceptibility to stress. Mol Psychiatry 25(5):951–964. https://doi.org/10.1038/s41380-019-0421-5
Li Z, Li X, Jin M, Liu Y, He Y, Jia N, Cui X, Liu Y et al (2022) Identification of potential biomarkers and their correlation with immune infiltration cells in schizophrenia using combinative bioinformatics strategy. Psychiatry Res 314:114658. https://doi.org/10.1016/j.psychres.2022.114658
Li Z, Li X, Jin M, Liu Y, He Y, Jia N, Cui X, Liu Y et al (2022) Identification of potential blood biomarkers for early diagnosis of schizophrenia through RNA sequencing analysis. J Psychiatr Res 147:39–49. https://doi.org/10.1016/j.jpsychires.2022.01.003
Xie M, Li Z, Li X, Ai L, Jin M, Jia N, Yang Y, Li W et al (2022) Identifying crucial biomarkers in peripheral blood of schizophrenia and screening therapeutic agents by comprehensive bioinformatics analysis. J Psychiatr Res 152:86–96. https://doi.org/10.1016/j.jpsychires.2022.06.007
Issler O, van der Zee YY, Ramakrishnan A, Xia S, Zinsmaier AK, Tan C, Li W, Browne CJ et al (2022) The long noncoding RNA FEDORA is a cell type- and sex-specific regulator of depression. Sci Adv 8(48):9494. https://doi.org/10.1126/sciadv.abn9494
Williams AV, Pena CJ, Ramos-Maciel S, Laman-Maharg A, Ordonez-Sanchez E, Britton M, Durbin-Johnson B, Settles M et al (2022) Comparative transcriptional analyses in the nucleus accumbens identifies RGS2 as a key mediator of depression-related behavior. Biol Psychiatry 92(12):942–951. https://doi.org/10.1016/j.biopsych.2022.06.030
Kolde R, Laur S, Adler P, Vilo J (2012) Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 28(4):573–580. https://doi.org/10.1093/bioinformatics/btr709
Mamdani F, Weber MD, Bunney B, Burke K, Cartagena P, Walsh D, Lee FS, Barchas J, Schatzberg AF, Myers RM, Watson SJ, Akil H, Vawter MP, Bunney WE, Sequeira A (2022) Identification of potential blood biomarkers associated with suicide in major depressive disorder. Transl Psychiatry 12(1):159. https://doi.org/10.1038/s41398-022-01918-w
Vazquez M, Nogales-Cadenas R, Arroyo J, Botias P, Garcia R, Carazo JM, Tirado F, Pascual-Montano A et al (2010) MARQ: an online tool to mine GEO for experiments with similar or opposite gene expression signatures. Nucleic Acids Res 38(Web Server issue):W228-232. https://doi.org/10.1093/nar/gkq476
van der Zee YY, Eijssen LMT, Mews P, Ramakrishnan A, Alvarez K, Lardner CK, Cates HM, Walker DM et al (2022) Blood miR-144-3p: a novel diagnostic and therapeutic tool for depression. Mol Psychiatry 27(11):4536–4549. https://doi.org/10.1038/s41380-022-01712-6
Xu K, Zheng P, Zhao S, Wang J, Feng J, Ren Y, Zhong Q, Zhang H et al (2023) LRFN5 and OLFM4 as novel potential biomarkers for major depressive disorder: a pilot study. Transl Psychiatry 13(1):188. https://doi.org/10.1038/s41398-023-02490-7
Choudary PV, Molnar M, Evans SJ, Tomita H, Li JZ, Vawter MP, Myers RM, Bunney WE Jr et al (2005) Altered cortical glutamatergic and GABAergic signal transmission with glial involvement in depression. Proc Natl Acad Sci USA 102(43):15653–15658. https://doi.org/10.1073/pnas.0507901102
Bernard R, Kerman IA, Thompson RC, Jones EG, Bunney WE, Barchas JD, Schatzberg AF, Myers RM et al (2011) Altered expression of glutamate signaling, growth factor, and glia genes in the locus coeruleus of patients with major depression. Mol Psychiatry 16(6):634–646. https://doi.org/10.1038/mp.2010.44
Chandley MJ, Szebeni K, Szebeni A, Crawford J, Stockmeier CA, Turecki G, Miguel-Hidalgo JJ, Ordway GA (2013) Gene expression deficits in pontine locus coeruleus astrocytes in men with major depressive disorder. J Psychiatry Neurosci 38(4):276–284. https://doi.org/10.1503/jpn.120110
Medina A, Burke S, Thompson RC, Bunney W Jr, Myers RM, Schatzberg A, Akil H, Watson SJ (2013) Glutamate transporters: a key piece in the glutamate puzzle of major depressive disorder. J Psychiatr Res 47(9):1150–1156. https://doi.org/10.1016/j.jpsychires.2013.04.007
Bhatt S, Nagappa AN, Patil CR (2020) Role of oxidative stress in depression. Drug Discov Today 25(7):1270–1276. https://doi.org/10.1016/j.drudis.2020.05.001
Bouvier E, Brouillard F, Molet J, Claverie D, Cabungcal JH, Cresto N, Doligez N, Rivat C et al (2017) Nrf2-dependent persistent oxidative stress results in stress-induced vulnerability to depression. Mol Psychiatry 22(12):1701–1713. https://doi.org/10.1038/mp.2016.144
Blizniewska-Kowalska K, Galecki P, Su KP, Halaris A, Szemraj J, Galecka M (2022) Expression of PON1, PON2, PON3 and MPO genes in patients with depressive disorders. J Clin Med 11 (12). https://doi.org/10.3390/jcm11123321
Rosu GC, Pirici I, Grigorie AA, Istrate-Ofiteru AM, Iovan L, Tudorica V, Pirici D (2019) Distribution of aquaporins 1 and 4 in the central nervous system. Curr Health Sci J 45(2):218–226. https://doi.org/10.12865/CHSJ.45.02.14
Luo Y, Zeng B, Zeng L, Du X, Li B, Huo R, Liu L, Wang H et al (2018) Gut microbiota regulates mouse behaviors through glucocorticoid receptor pathway genes in the hippocampus. Transl Psychiatry 8(1):187. https://doi.org/10.1038/s41398-018-0240-5
Hoshi A, Yamamoto T, Shimizu K, Ugawa Y, Nishizawa M, Takahashi H, Kakita A (2012) Characteristics of aquaporin expression surrounding senile plaques and cerebral amyloid angiopathy in Alzheimer disease. J Neuropathol Exp Neurol 71(8):750–759. https://doi.org/10.1097/NEN.0b013e3182632566
Balestri M, Crisafulli C, Donato L, Giegling I, Calati R, Antypa N, Schneider B, Marusic D et al (2017) Nine differentially expressed genes from a post mortem study and their association with suicidal status in a sample of suicide completers, attempters and controls. J Psychiatr Res 91:98–104. https://doi.org/10.1016/j.jpsychires.2017.03.009
Sokolowski M, Wasserman J, Wasserman D (2016) Polygenic associations of neurodevelopmental genes in suicide attempt. Mol Psychiatry 21(10):1381–1390. https://doi.org/10.1038/mp.2015.187
Kaut O, Sharma A, Schmitt I, Hurlemann R, Wullner U (2017) DNA methylation of DLG4 and GJA-1 of human hippocampus and prefrontal cortex in major depression is unchanged in comparison to healthy individuals. J Clin Neurosci 43:261–263. https://doi.org/10.1016/j.jocn.2017.05.030
Wu Z, Cai Z, Shi H, Huang X, Cai M, Yuan K, Huang P, Shi G et al (2022) Effective biomarkers and therapeutic targets of nerve-immunity interaction in the treatment of depression: an integrated investigation of the miRNA-mRNA regulatory networks. Aging (Albany NY) 14(8):3569–3596. https://doi.org/10.18632/aging.204030
Medina A, Watson SJ, Bunney W Jr, Myers RM, Schatzberg A, Barchas J, Akil H, Thompson RC (2016) Evidence for alterations of the glial syncytial function in major depressive disorder. J Psychiatr Res 72:15–21. https://doi.org/10.1016/j.jpsychires.2015.10.010
Liu T, Zhou N, Xu R, Cao Y, Zhang Y, Liu Z, Zheng X, Feng W (2020) A metabolomic study on the anti-depressive effects of two active components from Chrysanthemum morifolium. Artif Cells Nanomed Biotechnol 48(1):718–727. https://doi.org/10.1080/21691401.2020.1774597
Xie X, Yu C, Zhou J, Xiao Q, Shen Q, Xiong Z, Li Z, Fu Z (2020) Nicotinamide mononucleotide ameliorates the depression-like behaviors and is associated with attenuating the disruption of mitochondrial bioenergetics in depressed mice. J Affect Disord 263:166–174. https://doi.org/10.1016/j.jad.2019.11.147
Figaro-Drumond FV, Pereira SC, Menezes IC, von Werne BC, Coeli-Lacchini FB, Oliveira-Paula GH, Cleare AJ, Young AH et al (2020) Association of 11beta-hydroxysteroid dehydrogenase type1 (HSD11b1) gene polymorphisms with outcome of antidepressant therapy and suicide attempts. Behav Brain Res 381:112343. https://doi.org/10.1016/j.bbr.2019.112343
Mahmood N, Nawaz R, Kadir HA, Al Mughairbi F (2021) Genetic biomarkers in association with depressive disorder in UAE residents: a pilot case study. Oman Med J 36(4):e293. https://doi.org/10.5001/omj.2021.89
Banerjee S, Aykin-Burns N, Krager KJ, Shah SK, Melnyk SB, Hauer-Jensen M, Pawar SA (2016) Loss of C/EBPdelta enhances IR-induced cell death by promoting oxidative stress and mitochondrial dysfunction. Free Radic Biol Med 99:296–307. https://doi.org/10.1016/j.freeradbiomed.2016.08.022
Banerjee S, Alexander T, Majumdar D, Groves T, Kiffer F, Wang J, Gorantla A, Allen AR, et al (2019) Loss of C/EBPdelta exacerbates radiation-induced cognitive decline in aged mice due to impaired oxidative stress response. Int J Mol Sci 20 (4). https://doi.org/10.3390/ijms20040885
Austin SH, Harris RM, Booth AM, Lang AS, Farrar VS, Krause JS, Hallman TA, MacManes M et al (2021) Isolating the role of corticosterone in the hypothalamic-pituitary-gonadal transcriptomic stress response. Front Endocrinol (Lausanne) 12:632060. https://doi.org/10.3389/fendo.2021.632060
Suzuki K, Shibato J, Rakwal R, Takaura M, Hotta R, Masuo Y (2023) Biomarkers in the rat hippocampus and peripheral blood for an early stage of mental disorders induced by water immersion stress. Int J Mol Sci 24 (4). https://doi.org/10.3390/ijms24043153
Wang SM, Lim SW, Wang YH, Lin HY, Lai MD, Ko CY, Wang JM (2018) Astrocytic CCAAT/Enhancer-binding protein delta contributes to reactive oxygen species formation in neuroinflammation. Redox Biol 16:104–112. https://doi.org/10.1016/j.redox.2018.02.011
Acknowledgements
We would like to thank the participants involved in this study.
Funding
This study was supported by the Joint Project of Chongqing Municipal Science and Technology Bureau and Chongqing Health Commission (2023CCXM003), the Natural Science Foundation Project of China (81820108015, 82101596, and 82371526), the National Key Research and Development Program of China (2017YFA0505700), the Young Elite Scientists Sponsorship Program by CAST (2021QNRC001), the China Postdoctoral Science Foundation (2022MD723735), the Natural Science Foundation of Chongqing (cstc2022ycjh-bgzxm0033), and the Chongqing Postdoctoral Science Foundation (2022NSCQ-BHX1283).
Author information
Authors and Affiliations
Contributions
Conceptualization: Xiaogang Zhong, Yue Chen, Weiyi Chen, and Peng Xie; data curation: Siwen Gui: Juncai Pu and Dongfang Wang; formal analysis: Yiyun Liu and Juncai Pu; funding acquisition: Peng Xie; methodology: Xiaogang Zhong, Yue Chen, Weiyi Chen, and Yiyun Liu; software: Siwen Gui and Xiang Chen; supervision: Peng Xie; validation: Yue Chen, Yong He, Xiaopeng Chen, and Renjie Qiao; visualization: Xiaogang Zhong and Siwen Gui; writing—original draft: Xiaogang Zhong; writing—review and editing: Peng Xie.
Corresponding author
Ethics declarations
Ethics Approval
Human experimentation in this study was approved by the Ethics Committee of Chongqing Medical University (2017013).
Consent to Participate
For the publicly available data, the informed consent has been reported in the original studies. For the clinical validation cohort, all participants signed the informed consent.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhong, X., Chen, Y., Chen, W. et al. Identification of Potential Biomarkers for Major Depressive Disorder: Based on Integrated Bioinformatics and Clinical Validation. Mol Neurobiol (2024). https://doi.org/10.1007/s12035-024-04217-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12035-024-04217-1