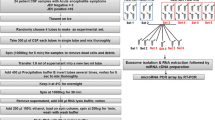
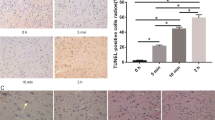
Abstract
Sepsis-associated encephalopathy (SAE) is a diffuse brain dysfunction secondary to body infection without overt central nervous system infection. Dysregulation of miRNA expression in the transcriptome can spread through RNA transfer in exosomes, providing an early signal of impending neuropathological changes in the brain. Here, we comprehensively analyzed brain-derived exosomal miRNA profiles in SAE rats (n = 3) and controls (n = 3). We further verified the differential expression and correlation of brain tissue, cerebrospinal fluid, and plasma exosomal miRNAs in SAE rats. High-throughput sequencing of brain-derived exosomal miRNAs identified 101 differentially expressed miRNAs, of which 16 were downregulated and 85 were upregulated. Four exosomal miRNAs (miR-127-3p, miR-423-3p, mR-378b, and miR-106-3p) were differentially expressed and correlated in the brain tissue, cerebrospinal fluid, and plasma, revealing the potential use of miRNAs as SAE liquid brain biopsies. Understanding exosomal miRNA profiles in SAE brain tissue and exploring the correlation with peripheral exosomal miRNA can contribute to a comprehensive understanding of miRNA changes in the SAE pathological process and provide the possibility of establishing early diagnostic assays.





Similar content being viewed by others
Data Availability
All relevant data for the present study are in the manuscript and Supplementary Information.
References
Gofton TE, Young GB (2012) Sepsis-associated encephalopathy. Nat Rev Neurol 8(10):557–566. https://doi.org/10.1038/nrneurol.2012.183
Ren C, Yao RQ, Zhang H, Feng YW, Yao YM (2020) Sepsis-associated encephalopathy: a vicious cycle of immunosuppression. J Neuroinflammation 17(1):14. https://doi.org/10.1186/s12974-020-1701-3
Helbing DL, Bohm L, Witte OW (2018) Sepsis-associated encephalopathy. CMAJ 190(36):E1083. https://doi.org/10.1503/cmaj.180454
Zhang LN, Wang XT, Ai YH, Guo QL, Huang L, Liu ZY, Yao B (2012) Epidemiological features and risk factors of sepsis-associated encephalopathy in intensive care unit patients: 2008-2011. Chin Med J 125(5):828–831
Cheng L, Vella LJ, Barnham KJ, McLean C, Masters CL, Hill AF (2020) Small RNA fingerprinting of Alzheimer’s disease frontal cortex extracellular vesicles and their comparison with peripheral extracellular vesicles. J Extracell Vesicles 9(1):1766822. https://doi.org/10.1080/20013078.2020.1766822
Kosik KS (2006) The neuronal microRNA system. Nat Rev Neurosci 7(12):911–920. https://doi.org/10.1038/nrn2037
Diaz NF, Cruz-Resendiz MS, Flores-Herrera H, Garcia-Lopez G, Molina-Hernandez A (2014) MicroRNAs in central nervous system development. Rev Neurosci 25(5):675–686. https://doi.org/10.1515/revneuro-2014-0014
Treiber T, Treiber N, Meister G (2019) Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol 20(1):5–20. https://doi.org/10.1038/s41580-018-0059-1
Ambros V (2001) MicroRNAs: tiny regulators with great potential. Cell 107(7):823–826. https://doi.org/10.1016/s0092-8674(01)00616-x
Rupaimoole R, Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16(3):203–222. https://doi.org/10.1038/nrd.2016.246
Ho PTB, Clark IM, Le LTT (2022) MicroRNA-based diagnosis and therapy. Int J Mol Sci 23(13):7167. https://doi.org/10.3390/ijms23137167
Xia X, Wang Y, Huang Y, Zhang H, Lu H, Zheng JC (2019) Exosomal miRNAs in central nervous system diseases: biomarkers, pathological mediators, protective factors and therapeutic agents. Prog Neurobiol 183:101694. https://doi.org/10.1016/j.pneurobio.2019.101694
Liang Y, Lehrich BM, Zheng S, Lu M (2021) Emerging methods in biomarker identification for extracellular vesicle-based liquid biopsy. J Extracell Vesicles 10(7):e12090. https://doi.org/10.1002/jev2.12090
Min L, Zhu S, Chen L, Liu X, Wei R, Zhao L, Yang Y et al. (2019) Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: a comparison with plasma total miRNAs. J Extracell Vesicles 8(1):1643670. https://doi.org/10.1080/20013078.2019.1643670
Kalluri R, LeBleu VS (2020) The biology, function, and biomedical applications of exosomes. Science 367(6478). https://doi.org/10.1126/science.aau6977
Jia L, Qiu Q, Zhang H, Chu L, Du Y, Zhang J, Zhou C, Liang F et al. (2019) Concordance between the assessment of Abeta42, T-tau, and P-T181-tau in peripheral blood neuronal-derived exosomes and cerebrospinal fluid. Alzheimers Dement 15(8):1071–1080. https://doi.org/10.1016/j.jalz.2019.05.002
Mittal R, Bencie N, Langlie J, Mittal J, Eshraghi AA (2021) Exosomes as drug delivery vehicles and biomarkers for neurological and auditory systems. J Cell Physiol 236(12):8035–8049. https://doi.org/10.1002/jcp.30484
Wood MJ, O'Loughlin AJ, Samira L (2011) Exosomes and the blood-brain barrier: implications for neurological diseases. Ther Deliv 2(9):1095–1099. https://doi.org/10.4155/tde.11.83
Bub A, Brenna S, Alawi M, Kugler P, Gui Y, Kretz O, Altmeppen H, Magnus T et al. (2022) Multiplexed mRNA analysis of brain-derived extracellular vesicles upon experimental stroke in mice reveals increased mRNA content with potential relevance to inflammation and recovery processes. Cell Mol Life Sci 79(6):329. https://doi.org/10.1007/s00018-022-04357-4
Wei X-b, Jiang W-q, Zeng J-h, Huang L-q, Ding H-g, Jing Y-w, Han Y-l, Li Y-c et al. (2022) Exosome-derived lncRNA NEAT1 exacerbates sepsis-associated encephalopathy by promoting ferroptosis through regulating miR-9-5p/TFRC and GOT1 axis. Mol Neurobiol 59(3):1954–1969. https://doi.org/10.1007/s12035-022-02738-1
Kikuchi DS, Campos ACP, Qu H, Forrester SJ, Pagano RL, Lassegue B, Sadikot RT, Griendling KK, Hernandes MS (2019) Poldip2 mediates blood-brain barrier disruption in a model of sepsis-associated encephalopathy. J Neuroinflammation 16(1):241. https://doi.org/10.1186/s12974-019-1575-4
Siami S, Annane D, Sharshar T (2008) The encephalopathy in sepsis. Crit Care Clin 24(1):67–82, viii. https://doi.org/10.1016/j.ccc.2007.10.001
Song G, Liang H, Song H, Ding X, Wang D, Zhang X, Sun T (2022) Metformin improves the prognosis of adult mice with sepsis-associated encephalopathy better than that of aged mice. J Immunol Res 2022:3218452. https://doi.org/10.1155/2022/3218452
Kafa IM, Uysal M, Bakirci S, Ayberk Kurt M (2010) Sepsis induces apoptotic cell death in different regions of the brain in a rat model of sepsis. Acta Neurobiol Exp 70(3):246–260
Radu M, Chernoff J (2013) An in vivo assay to test blood vessel permeability. J Vis Exp 73:e50062. https://doi.org/10.3791/50062
Xue X, Shen YL, Cheng YY, Rong J, Zhou H, Chi H (2021) 0D/2D coordination complexes: magnetic studies, protection, and mechanism against ischemic myocardial damage by reducing the activation of the PI3K/Akt signaling pathway. ACS Omega 6(8):5408–5414. https://doi.org/10.1021/acsomega.0c05601
Vella LJ, Scicluna BJ, Cheng L, Bawden EG, Masters CL, Ang CS, Willamson N, McLean C, Barnham KJ, Hill AF (2017) A rigorous method to enrich for exosomes from brain tissue. J Extracell Vesicles 6(1):1348885. https://doi.org/10.1080/20013078.2017.1348885
Tian Y, Gong M, Hu Y, Liu H, Zhang W, Zhang M, Hu X, Aubert D et al. (2020) Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J Extracell Vesicles 9(1):1697028. https://doi.org/10.1080/20013078.2019.1697028
Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV et al. (2014) Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 3:26913. https://doi.org/10.3402/jev.v3.26913
Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N (2012) miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40(1):37–52. https://doi.org/10.1093/nar/gkr688
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. https://doi.org/10.1093/bioinformatics/btp616
Betel D, Wilson M, Gabow A, Marks DS, Sander C (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res 36(Database issue):D149–D153. https://doi.org/10.1093/nar/gkm995
Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10(10):1507–1517. https://doi.org/10.1261/rna.5248604
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25(1):25–29. https://doi.org/10.1038/75556
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32(Database issue):D277–D280. https://doi.org/10.1093/nar/gkh063
Niu M, Li H, Li X, Yan X, Ma A, Pan X, Zhu X (2021) Circulating exosomal miRNAs as novel biomarkers perform superior diagnostic efficiency compared with plasma miRNAs for large-artery atherosclerosis stroke. Front Pharmacol 12:791644. https://doi.org/10.3389/fphar.2021.791644
Yan X, Yang K, Xiao Q, Hou R, Pan X, Zhu X (2022) Central role of microglia in sepsis-associated encephalopathy: from mechanism to therapy. Front Immunol 13:929316. https://doi.org/10.3389/fimmu.2022.929316
van Niel G, D'Angelo G, Raposo G (2018) Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19(4):213–228. https://doi.org/10.1038/nrm.2017.125
Upadhya R, Zingg W, Shetty S, Shetty AK (2020) Astrocyte-derived extracellular vesicles: neuroreparative properties and role in the pathogenesis of neurodegenerative disorders. J Control Release 323:225–239. https://doi.org/10.1016/j.jconrel.2020.04.017
Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5(3):R13. https://doi.org/10.1186/gb-2004-5-3-r13
Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR (2004) Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol 5(9):R68. https://doi.org/10.1186/gb-2004-5-9-r68
Kim YJ, Yeon Y, Lee WJ, Shin YU, Cho H, Lim HW, Kang MH (2022) Analysis of microRNA expression in tears of patients with herpes epithelial keratitis: a preliminary study. Investigative Opthalmology & Visual. Science 63(4):21. https://doi.org/10.1167/iovs.63.4.21
Yang YH, Zhu J (2019) Targeting miR-106-3p facilitates functional recovery via inactivating inflammatory microglia and interfering glial scar component deposition after neural injury. Eur Rev Med Pharmacol Sci 23(20):9000–9008. https://doi.org/10.26355/eurrev_201910_19300
Lundy SR, Abney K, Ellerson D, Igietseme JU, Carroll D, Eko FO, Omosun YO (2021) MiR-378b modulates chlamydia-induced upper genital tract pathology. Pathogens 10(5):566. https://doi.org/10.3390/pathogens10050566
Link F, Krohn K, Schumann J (2019) Identification of stably expressed housekeeping miRNAs in endothelial cells and macrophages in an inflammatory setting. Sci Rep 9(1):12786. https://doi.org/10.1038/s41598-019-49241-7
Zhang ZB, Xiong LL, Xue LL, Deng YP, Du RL, Hu Q, Xu Y, Yang SJ et al. (2021) MiR-127-3p targeting CISD1 regulates autophagy in hypoxic-ischemic cortex. Cell Death Dis 12(3):279. https://doi.org/10.1038/s41419-021-03541-x
Challagundla KB, Fanini F, Vannini I, Wise P, Murtadha M, Malinconico L, Cimmino A, Fabbri M (2014) MicroRNAs in the tumor microenvironment: solving the riddle for a better diagnostics. Expert Rev Mol Diagn 14(5):565–574. https://doi.org/10.1586/14737159.2014.922879
Thind A, Wilson C (2016) Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles 5:31292. https://doi.org/10.3402/jev.v5.31292
Koga Y, Yasunaga M, Moriya Y, Akasu T, Fujita S, Yamamoto S, Matsumura Y (2011) Exosome can prevent RNase from degrading microRNA in feces. J Gastrointest Oncol 2(4):215–222. https://doi.org/10.3978/j.issn.2078-6891.2011.015
Bebelman MP, Smit MJ, Pegtel DM, Baglio SR (2018) Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther 188:1–11. https://doi.org/10.1016/j.pharmthera.2018.02.013
Fiandaca MS, Kapogiannis D, Mapstone M, Boxer A, Eitan E, Schwartz JB, Abner EL, Petersen RC et al. (2015) Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: a case-control study. Alzheimers Dement 11(6):600–607.e601. https://doi.org/10.1016/j.jalz.2014.06.008
Acknowledgements
We thank the Institute of Cerebrovascular Diseases and Medical Research Center of the Affiliated Hospital of Qingdao University for the platform support.
Funding
This work was supported by the Natural Science Foundation of Shandong Province (ZR2020MH138).
Author information
Authors and Affiliations
Contributions
Q.X., X.Y., R.H., X.P., and X.Z. contributed to the conception and design of the study; Q.X., X.Y., Y.S., and Y.T. performed the experiments and data collection and analyzed the data; Q.X., X.Y., and X.Z. contributed to drafting the text or preparing the figures.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
All animal experiments were approved by the Experimental Animal Management Committee of the Affiliated Hospital of Qingdao University.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 479 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiao, Q., Yan, X., Sun, Y. et al. Brain-Derived Exosomal miRNA Profiles upon Experimental SAE Rats and Their Comparison with Peripheral Exosomes. Mol Neurobiol 61, 772–782 (2024). https://doi.org/10.1007/s12035-023-03569-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-023-03569-4