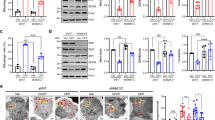
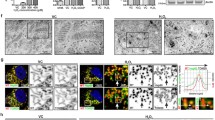
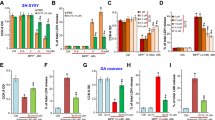
Abstract
Sterile α and toll/interleukin 1 receptor motif-containing protein 1 (SARM1) is the defining molecule and central executioner of programmed axon death, also known as Wallerian degeneration. SARM1 has a mitochondrial targeting sequence, and it can bind to and stabilize PTEN-induced putative kinase 1 (PINK1) for mitophagy induction, but the deletion of the mitochondrial localization sequence is found to disrupt the mitochondrial localization of SARM1 in neurons without altering its ability to promote axon degeneration after axotomy. The biological significance of SARM1 mitochondrial localization remains elusive. In this study, we observed that the pro-degeneration factor, SARM1, was upregulated in acrylamide (ACR) neuropathy, a slow, Wallerian-like, programmed axonal death process. The upregulated SARM1 accumulated on mitochondria, interfered with mitochondrial dynamics, and activated PINK1-mediated mitophagy. Importantly, rapamycin (RAPA) intervention eliminated mitochondrial accumulation of SARM1 and partly attenuated ACR neuropathy. Thus, mitochondrial localization of SARM1 may contribute to its clearance through the SARM1-PINK1 mitophagy pathway, which inhibits axonal degeneration through a negative feedback loop. The mitochondrial localization of SARM1 complements the coordinated activity of the pro-survival factor, nicotinamide mononucleotide adenyltransferase 2 (NMNAT2), and SARM1 and is part of the self-limiting molecular mechanisms underpinning programmed axon death in ACR neuropathy.
Graphical abstract
Mitophagy clearance of SARM1 is complementary to the coordinated activity of NMNAT2 and SARM1 in ACR neuropathy.






Similar content being viewed by others
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ACR :
-
Acrylamide
- ARM :
-
Armadillo repeats
- cADPR :
-
Cyclic adenosine diphosphate ribose
- NAD + :
-
Nicotinamide adenine dinucleotide
- NMN :
-
Nicotinamide mononucleotide
- NMNAT2 :
-
Nicotinamide mononucleotide adenyltransferase 2
- PINK1 :
-
PTEN-induced putative kinase 1
- RAPA :
-
Rapamycin
- SAM :
-
Sterile α motif
- SARM :
-
Sterile α and toll/interleukin 1 receptor motif-containing protein 1
- TIR :
-
Toll/interleukin 1 receptor
- TRAF6 :
-
Tumor necrosis factor receptor-associated factor 6
References
Waller A (1851) Experiments on the section of the glosso-pharyngeal and hypoglossal nerves of the frog, and observations of the alterations produced thereby in the structure of their primitive fibres. Edinburgh Med Surg J 76(189):369–376
Perry VH, Brown MC, Lunn ER, Tree P, Gordon S (1990) Evidence that very slow Wallerian degeneration in C57BL/Ola mice is an intrinsic property of the peripheral nerve. Eur J Neurosci 2(9):802–808. https://doi.org/10.1111/j.1460-9568.1990.tb00472.x
Perry VH, Brown MC, Lunn ER (1991) Very slow retrograde and Wallerian degeneration in the CNS of C57BL/Ola mice. Eur J Neurosci 3(1):102–105. https://doi.org/10.1111/j.1460-9568.1991.tb00815.x
Lyon MF, Ogunkolade BW, Brown MC, Atherton DJ, Perry VH (1993) A gene affecting Wallerian nerve degeneration maps distally on mouse chromosome 4. Proc Natl Acad Sci USA 90(20):9717–9720. https://doi.org/10.1073/pnas.90.20.9717
Desbois M, Crawley O, Evans PR, Baker ST, Masuho I, Yasuda R, Grill B (2018) PAM forms an atypical SCF ubiquitin ligase complex that ubiquitinates and degrades NMNAT2. J Biol Chem 293(36):13897–13909. https://doi.org/10.1074/jbc.RA118.002176
Babetto E, Beirowski B, Russler EV, Milbrandt J, DiAntonio A (2013) The Phr1 ubiquitin ligase promotes injury-induced axon self-destruction. Cell Rep 3(5):1422–1429. https://doi.org/10.1016/j.celrep.2013.04.013
Essuman K, Summers DW, Sasaki Y, Mao X, DiAntonio A, Milbrandt J (2017) The SARM1 toll/interleukin-1 receptor domain possesses intrinsic NAD(+) cleavage activity that promotes pathological axonal degeneration. Neuron 93(6):1334-1343.e1335. https://doi.org/10.1016/j.neuron.2017.02.022
Gerdts J, Summers DW, Milbrandt J, DiAntonio A (2016) Axon self-destruction: new links among SARM1, MAPKs, and NAD+ metabolism. Neuron 89(3):449–460. https://doi.org/10.1016/j.neuron.2015.12.023
Mink M, Fogelgren B, Olszewski K, Maroy P, Csiszar K (2001) A novel human gene (SARM) at chromosome 17q11 encodes a protein with a SAM motif and structural similarity to Armadillo/beta-catenin that is conserved in mouse, Drosophila, and Caenorhabditis elegans. Genomics 74(2):234–244. https://doi.org/10.1006/geno.2001.6548
Meijer AH, Gabby Krens SF, Medina Rodriguez IA, He S, Bitter W, Ewa Snaar-Jagalska B, Spaink HP (2004) Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol Immunol 40(11):773–783. https://doi.org/10.1016/j.molimm.2003.10.003
Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ (2004) TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol 5(5):488–494. https://doi.org/10.1038/ni1060
Belinda LW, Wei WX, Hanh BT, Lei LX, Bow H, Ling DJ (2008) SARM: a novel Toll-like receptor adaptor, is functionally conserved from arthropod to human. Mol Immunol 45(6):1732–1742. https://doi.org/10.1016/j.molimm.2007.09.030
Panneerselvam P, Singh Laishram P, Ho B, Chen J, Ding Jeak L (2012) Targeting of pro-apoptotic TLR adaptor SARM to mitochondria: definition of the critical region and residues in the signal sequence. Biochem J 442(2):263–271. https://doi.org/10.1042/BJ20111653%JBiochemicalJournal
Murata H, Sakaguchi M, Kataoka K, Huh N-H (2013) SARM1 and TRAF6 bind to and stabilize PINK1 on depolarized mitochondria. Mol Biol Cell 24(18):2772–2784. https://doi.org/10.1091/mbc.E13-01-0016
Loreto A, Di Stefano M, Gering M, Conforti L (2015) Wallerian degeneration is executed by an NMN-SARM1-dependent late Ca(2+) influx but only modestly influenced by mitochondria. Cell Rep 13(11):2539–2552. https://doi.org/10.1016/j.celrep.2015.11.032
Gerdts J, Summers DW, Sasaki Y, DiAntonio A, Milbrandt J (2013) Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J Neurosci 33(33):13569–13580. https://doi.org/10.1523/jneurosci.1197-13.2013
Pennisi M, Malaguarnera G, Puglisi V, Vinciguerra L, Vacante M, Malaguarnera M (2013) Neurotoxicity of acrylamide in exposed workers. Int J Environ Res Public Health 10(9):3843–3854. https://doi.org/10.3390/ijerph10093843
Tareke E, Rydberg P, Karlsson P, Eriksson S, Törnqvist M (2002) Analysis of acrylamide, a carcinogen formed in heated foodstuffs. J Agric Food Chem 50(17):4998–5006. https://doi.org/10.1021/jf020302f
Schettgen T, Weiss T, Drexler H, Angerer J (2003) A first approach to estimate the internal exposure to acrylamide in smoking and non-smoking adults from Germany. Int J Hyg Environ Health 206(1):9–14. https://doi.org/10.1078/1438-4639-00195
Urban M, Kavvadias D, Riedel K, Scherer G, Tricker AR (2006) Urinary mercapturic acids and a hemoglobin adduct for the dosimetry of acrylamide exposure in smokers and nonsmokers. Inhal Toxicol 18(10):831–839. https://doi.org/10.1080/08958370600748430
Lantz I, Ternité R, Wilkens J, Hoenicke K, Guenther H, van der Stegen GH (2006) Studies on acrylamide levels in roasting, storage and brewing of coffee. Mol Nutr Food Res 50(11):1039–1046. https://doi.org/10.1002/mnfr.200600069
He FS, Zhang SL, Wang HL, Li G, Zhang ZM, Li FL, Dong XM, Hu FR (1989) Neurological and electroneuromyographic assessment of the adverse effects of acrylamide on occupationally exposed workers. Scand J Work Environ Health 15(2):125–129. https://doi.org/10.5271/sjweh.1878
Deng H, He F, Zhang S, Calleman CJ, Costa LG (1993) Quantitative measurements of vibration threshold in healthy adults and acrylamide workers. Int Arch Occup Environ Health 65(1):53–56. https://doi.org/10.1007/bf00586059
Edwards PM, Parker VH (1977) A simple, sensitive, and objective method for early assessment of acrylamide neuropathy in rats. Toxicol Appl Pharmacol 40(3):589–591. https://doi.org/10.1016/0041-008x(77)90083-7
Kuperman AS (1958) Effects of acrylamide on the central nervous system of the cat. J Pharmacol Exp Ther 123(3):180–192
Fullerton PM, Barnes JM (1966) Peripheral neuropathy in rats produced by acrylamide. Br J Ind Med 23(3):210–221. https://doi.org/10.1136/oem.23.3.210
Hopkins A (1970) The effect of acrylamide on the peripheral nervous system of the baboon. J Neurol Neurosurg Psychiatry 33(6):805–816. https://doi.org/10.1136/jnnp.33.6.805
Hopkins AP, Gilliatt RW (1971) Motor and sensory nerve conduction velocity in the baboon: normal values and changes during acrylamide neuropathy. J Neurol Neurosurg Psychiatry 34(4):415–426. https://doi.org/10.1136/jnnp.34.4.415
Sumner AJ, Asbury AK (1974) Acrylamide neuropathy: selective vulnerability of sensory fibers. Trans Am Neurol Assoc 99:79–83
Sumner AJ, Asbury AK (1975) Physiological studies of the dying-back phenomenon. Muscle stretch afferents in acrylamide neuropathy. Brain 98(1):91–100. https://doi.org/10.1093/brain/98.1.91-a
Erkekoglu P, Baydar T (2014) Acrylamide neurotoxicity. Nutr Neurosci 17(2):49–57. https://doi.org/10.1179/1476830513y.0000000065
LoPachin RM, Barber DS, He D, Das S (2006) Acrylamide inhibits dopamine uptake in rat striatal synaptic vesicles. Toxicol Sci 89(1):224–234. https://doi.org/10.1093/toxsci/kfj005
Barber DS, Stevens S, LoPachin RM (2007) Proteomic analysis of rat striatal synaptosomes during acrylamide intoxication at a low dose rate. Toxicol Sci 100(1):156–167. https://doi.org/10.1093/toxsci/kfm210
Ogawa B, Ohishi T, Wang L, Takahashi M, Taniai E, Hayashi H, Mitsumori K, Shibutani M (2011) Disruptive neuronal development by acrylamide in the hippocampal dentate hilus after developmental exposure in rats. Arch Toxicol 85(8):987–994. https://doi.org/10.1007/s00204-010-0622-9
Crowe A, Bruelisauer A, Duerr L, Guntz P, Lemaire M (1999) Absorption and intestinal metabolism of SDZ-RAD and rapamycin in rats. Drug Metab Dispos 27(5):627–632
Rapamycin confers neuroprotection against aging-induced oxidative stress, mitochondrial dysfunction, and neurodegeneration in old rats through activation of autophagy (2019). 22 (1):60–70. https://doi.org/10.1089/rej.2018.2070
Hadamitzky M, Herring A, Kirchhof J, Bendix I, Haight MJ, Keyvani K, Lückemann L, Unteroberdörster M, Schedlowski M (2018) Repeated systemic treatment with rapamycin affects behavior and amygdala protein expression in rats. Int J Neuropsychopharmacol 21(6):592–602. https://doi.org/10.1093/ijnp/pyy017%
Clifton GL, Jiang JY, Lyeth BG, Jenkins LW, Hamm RJ, Hayes RL (1991) Marked protection by moderate hypothermia after experimental traumatic brain injury. J Cerebral Blood Flow Metabolism : Off J Int Soc Cerebral Blood Flow Metabolism 11(1):114–121. https://doi.org/10.1038/jcbfm.1991.13
Monville C, Torres EM, Dunnett SB (2006) Comparison of incremental and accelerating protocols of the rotarod test for the assessment of motor deficits in the 6-OHDA model. J Neurosci Methods 158(2):219–223. https://doi.org/10.1016/j.jneumeth.2006.06.001
Youssef AF, Santi BW (1997) Simple neurobehavioral functional observational battery and objective gait analysis validation by the use of acrylamide and methanol with a built-in recovery period. Environ Res 73(1):52–62. https://doi.org/10.1006/enrs.1997.3718
Moser VC, Anthony DC, Sette WF, MacPhail RC (1992) Comparison of subchronic neurotoxicity of 2-hydroxyethyl acrylate and acrylamide in rats. Fundam Appl Toxicol 18(3):343–352. https://doi.org/10.1016/0272-0590(92)90132-2
LoPachin RM, Ross JF, Reid ML, Das S, Mansukhani S, Lehning EJ (2002) Neurological evaluation of toxic axonopathies in rats: acrylamide and 2,5-hexanedione. Neurotoxicology 23(1):95–110. https://doi.org/10.1016/s0161-813x(02)00003-7
Takeuchi Y, Ono Y, Hisanaga N, Kitoh J, Sugiura Y (1980) A comparative study on the neurotoxicity of n-pentane, n-hexane, and n-heptane in the rat. Br J Ind Med 37(3):241–247. https://doi.org/10.1136/oem.37.3.241
Lima RT, Sousa D, Paiva AM, Palmeira A, Barbosa J, Pedro M, Pinto MM, Sousa E, Vasconcelos MH (2016) Modulation of autophagy by a thioxanthone decreases the viability of melanoma cells. Molecules (Basel, Switzerland) 21 (10). https://doi.org/10.3390/molecules21101343
Kong FJ, Wu JH, Sun SY, Zhou JQ (2017) The endoplasmic reticulum stress/autophagy pathway is involved in cholesterol-induced pancreatic β-cell injury. Sci Rep 7:44746. https://doi.org/10.1038/srep44746
Burek JD, Albee RR, Beyer JE, Bell TJ, Carreon RM, Morden DC, Wade CE, Hermann EA, Gorzinski SJ (1980) Subchronic toxicity of acrylamide administered to rats in the drinking water followed by up to 144 days of recovery. J Environ Pathol Toxicol 4(5–6):157–182
Hayakawa T, Kato K, Hayakawa R, Hisamoto N, Matsumoto K, Takeda K, Ichijo H (2011) Regulation of anoxic death in Caenorhabditis elegans by mammalian apoptosis signal-regulating kinase (ASK) family proteins. Genetics 187(3):785–792. https://doi.org/10.1534/genetics.110.124883
Kim Y, Zhou P, Qian L, Chuang J-Z, Lee J, Li C, Iadecola C, Nathan C, Ding A (2007) MyD88-5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. J Exp Med 204(9):2063–2074. https://doi.org/10.1084/jem.20070868
Mukherjee P, Winkler CW, Taylor KG, Woods TA, Nair V, Khan BA, Peterson KE (2015) SARM1, not MyD88, mediates TLR7/TLR9-induced apoptosis in neurons. J Immunol (Baltimore, Md : 1950) 195(10):4913–4921. https://doi.org/10.4049/jimmunol.1500953
Killackey SA, Rahman MA, Soares F, Zhang AB, Abdel-Nour M, Philpott DJ, Girardin SE (2019) The mitochondrial Nod-like receptor NLRX1 modifies apoptosis through SARM1. Mol Cell Biochem 453(1):187–196. https://doi.org/10.1007/s11010-018-3444-3
Geisler S, Doan RA, Cheng GC, Cetinkaya-Fisgin A, Huang SX, Höke A, Milbrandt J, DiAntonio A (2019) Vincristine and bortezomib use distinct upstream mechanisms to activate a common SARM1-dependent axon degeneration program. JCI Insight 4(17):e129920. https://doi.org/10.1172/jci.insight.129920
Bosanac T, Hughes RO, Engber T, Devraj R, Brearley A, Danker K, Young K, Kopatz J, Hermann M, Berthemy A, Boyce S, Bentley J, Krauss R (2021) Pharmacological SARM1 inhibition protects axon structure and function in paclitaxel-induced peripheral neuropathy. Brain. https://doi.org/10.1093/brain/awab184
Prats E, Gómez-Canela C, Ben-Lulu S, Ziv T, Padrós F, Tornero D, Garcia-Reyero N, Tauler R, Admon A, Raldúa D (2017) Modelling acrylamide acute neurotoxicity in zebrafish larvae. Sci Rep 7(1):13952. https://doi.org/10.1038/s41598-017-14460-3
MacAskill AF, Kittler JT (2010) Control of mitochondrial transport and localization in neurons. Trends Cell Biol 20(2):102–112. https://doi.org/10.1016/j.tcb.2009.11.002
Chen H, Chan DC (2009) Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum Mol Genet 18(R2):R169-176. https://doi.org/10.1093/hmg/ddp326
Schon EA, Przedborski S (2011) Mitochondria: the next (neurode)generation. Neuron 70(6):1033–1053. https://doi.org/10.1016/j.neuron.2011.06.003
Chan DC (2006) Mitochondria: dynamic organelles in disease, aging, and development. Cell 125(7):1241–1252. https://doi.org/10.1016/j.cell.2006.06.010
Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC (2004) Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet 36(6):585–595. https://doi.org/10.1038/ng1362
Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T (2008) The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest 118(6):2190–2199. https://doi.org/10.1172/jci33585
Yang Y, Chen S, Zhang J, Li C, Sun Y, Zhang L, Zheng X (2014) Stimulation of autophagy prevents amyloid-β peptide-induced neuritic degeneration in PC12 cells. Journal of Alzheimer’s disease : JAD 40(4):929–939. https://doi.org/10.3233/jad-132270
Clarke JP, Mearow K (2016) Autophagy inhibition in endogenous and nutrient-deprived conditions reduces dorsal root ganglia neuron survival and neurite growth in vitro. J Neurosci Res 94(7):653–670. https://doi.org/10.1002/jnr.23733
Wang Y, Song M, Song F (2018) Neuronal autophagy and axon degeneration. Cell Mol Life Sci 75(13):2389–2406. https://doi.org/10.1007/s00018-018-2812-1
Kim I, Rodriguez-Enriquez S, Lemasters JJ (2007) Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 462(2):245–253. https://doi.org/10.1016/j.abb.2007.03.034
Han S, Jeong YY, Sheshadri P, Su X, Cai Q (2020) Mitophagy regulates integrity of mitochondria at synapses and is critical for synaptic maintenance. EMBO reports 21(9):e49801. https://doi.org/10.15252/embr.201949801
Kim I, Lemasters JJ (2011) Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxid Redox Signal 14(10):1919–1928. https://doi.org/10.1089/ars.2010.3768
Pollock L, Jardine J, Urbé S, Clague MJ (2021) The PINK1 repertoire: not just a one trick pony. BioEssays 43(11):e2100168. https://doi.org/10.1002/bies.202100168
Loring HS, Parelkar SS, Mondal S, Thompson PR (2020) Identification of the first noncompetitive SARM1 inhibitors. Bioorg Med Chem 28(18):115644. https://doi.org/10.1016/j.bmc.2020.115644
Hughes RO, Bosanac T, Mao X, Engber TM, DiAntonio A, Milbrandt J, Devraj R, Krauss R (2021) Small molecule SARM1 inhibitors recapitulate the SARM1−/− phenotype and allow recovery of a metastable pool of axons fated to degenerate. Cell Rep 34(1):108588. https://doi.org/10.1016/j.celrep.2020.108588
Bosanac T, Hughes RO, Engber T, Devraj R, Brearley A, Danker K, Young K, Kopatz J, Hermann M, Berthemy A, Boyce S, Bentley J, Krauss R (2021) Pharmacological SARM1 inhibition protects axon structure and function in paclitaxel-induced peripheral neuropathy. Brain 144(10):3226–3238. https://doi.org/10.1093/brain/awab184
Li WH, Huang K, Cai Y, Wang QW, Zhu WJ, Hou YN, Wang S, Cao S, Zhao ZY, Xie XJ, Du Y, Lee CS, Lee HC, Zhang H, Zhao YJ (2021) Permeant fluorescent probes visualize the activation of SARM1 and uncover an anti-neurodegenerative drug candidate. eLife 10. https://doi.org/10.7554/eLife.67381
Figley MD, Gu W, Nanson JD, Shi Y, Sasaki Y, Cunnea K, Malde AK, Jia X, Luo Z, Saikot FK, Mosaiab T, Masic V, Holt S, Hartley-Tassell L, McGuinness HY, Manik MK, Bosanac T, Landsberg MJ, Kerry PS, Mobli M, Hughes RO, Milbrandt J, Kobe B, DiAntonio A, Ve T (2021) SARM1 is a metabolic sensor activated by an increased NMN/NAD+ ratio to trigger axon degeneration. Neuron. https://doi.org/10.1016/j.neuron.2021.02.009
Sasaki Y, Engber TM, Hughes RO, Figley MD, Wu T, Bosanac T, Devraj R, Milbrandt J, Krauss R, DiAntonio A (2020) cADPR is a gene dosage-sensitive biomarker of SARM1 activity in healthy, compromised, and degenerating axons. Exp Neurol 329:113252. https://doi.org/10.1016/j.expneurol.2020.113252
Zhang C, Kang K, Chen Y, Shan S, Xie K, Song F (2021) Atg7 knockout alleviated the axonal injury of neuro-2a cells induced by tri-ortho-cresyl phosphate. Neurotox Res 39(4):1076–1086. https://doi.org/10.1007/s12640-021-00344-y
Sur M, Dey P, Sarkar A, Bar S, Banerjee D, Bhat S, Mukherjee P (2018) Sarm1 induction and accompanying inflammatory response mediates age-dependent susceptibility to rotenone-induced neurotoxicity. Cell death discovery 4:114. https://doi.org/10.1038/s41420-018-0119-5
Acknowledgements
The authors want to thank the Translational Medicine Core Facility of Shandong University for consultation and instrument availability that supported this work, Editage (www.editage.cn) for English language editing, Lihua Yu and Wenwen Zheng for their assistance in experiments, and Wenhao Yu and Ziqiang Yu for their generous help in data analysis.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82173552; No. 81673209).
Author information
Authors and Affiliations
Contributions
Shuai Wang, a major contributor in writing the manuscript, performed the examination and analyzed and interpreted the data. Mingxue Song, Hui Yong, Cuiqin Zhang, Kang Kang, Zhidan Liu, Yiyu Yang, Zhengcheng Huang, Shu’e Wang Haotong Ge, and Xiulan Zhao contributed to the acquisition of data. Fuyong Song was responsible for the conception and design of the work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Animals were treated according to the NIH Guide for Care and Use of Laboratory Animals and followed the principles in the “Use of Animals in Toxicology.” All protocols were approved by the Institutional Animal Care and Use Committee of Shandong University.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• The pro-degeneration factor, SARM1, is upregulated in ACR neuropathy.
• The upregulated SARM1 accumulates on mitochondria during ACR neuropathy.
• ACR induces mitophagy, and the SARM1-PINK1 pathway is one of the underlying molecular mechanisms.
• RAPA intervention clears mitochondrial SARM1 and partly attenuates ACR neuropathy.
• Mitophagy clearance of SARM1 complements the coordinated activity of NMNAT2 and SARM1.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, S., Song, M., Yong, H. et al. Mitochondrial Localization of SARM1 in Acrylamide Intoxication Induces Mitophagy and Limits Neuropathy. Mol Neurobiol 59, 7337–7353 (2022). https://doi.org/10.1007/s12035-022-03050-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-03050-8