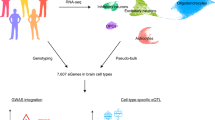
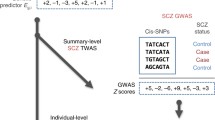
Abstract
Genome-wide association studies (GWAS) have identified a large number of schizophrenia risk variants, and most of them are mapped to noncoding regions. By leveraging multiple joint-tissue gene expression data and GWAS data, we herein performed a transcriptome-wide association study (TWAS) and Mendelian randomization (MR) analysis and identified 144 genes whose mRNA levels were related to genetic risk of schizophrenia. Most of these genes exhibited diametrically opposite trends of expression in prenatal and postnatal brain tissues, despite that their expression levels in dorsolateral prefrontal cortex (DLPFC) tissues did not significantly differ between schizophrenics and healthy controls. We then found significant enrichment of these genes in dopamine-related pathways that were repeatedly implicated in schizophrenia pathogenesis and in the action of antipsychotic drugs. Gene expression analysis using single cell RNA-sequencing (scRNA-seq) data of mid-gestation fetal brains further revealed enrichment of these genes in glutamatergic excitatory neurons and cycling progenitors. These lines of evidence, in consistency with previous findings, confirmed the polygenic nature of schizophrenia and highlighted involvement of early neurodevelopment aberrations in this disorder. Further investigations using advanced algorithms in both bulk brain tissues and in single cells and at different developmental stages are necessary to characterize transcriptomic features of schizophrenia pathogenesis along brain development.





Similar content being viewed by others
Data Availability
All data generated or analyzed in this study are included in the current manuscript and supplementary materials.
Code Availability
Not applicable.
References
Marder SR, Cannon TD (2019) Schizophrenia. N Engl J Med 381(18):1753–1761
Pardinas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S et al (2018) Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 50(3):381–389
Lam M, Chen CY, Li Z, Martin AR, Bryois J, Ma X, Gaspar H, Ikeda M et al (2019) Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet 51(12):1670–1678
Schizophrenia Working Group of the Psychiatric Genomics Consortium (2020) Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. medRxiv https://doi.org/10.1101/2020.09.12.20192922.
Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature 511(7510):421–427
Sullivan PF, Geschwind DH (2019) Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric Disorders. Cell 177(1):162–183
Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC et al (2016) Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 19(11):1442–1453
Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ (2010) Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 6 (4):e1000888
Ward LD, Kellis M (2012) Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol 30(11):1095–1106
Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R et al (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science 337(6099):1190–1195
Wang JY, Li XY, Li HJ, Liu JW, Yao YG, Li M, Xiao X, Luo XJ (2021) Integrative analyses followed by functional characterization reveal TMEM180 as a schizophrenia risk gene. Schizophr Bull. https://doi.org/10.1093/schbul/sbab032
Wu Y, Bi R, Zeng C, Ma C, Sun C, Li J, Xiao X, Li M et al (2019) Identification of the primate-specific gene BTN3A2 as an additional schizophrenia risk gene in the MHC loci. EBioMedicine 44:530–541
Huckins LM, Dobbyn A, Ruderfer DM, Hoffman G, Wang W, Pardinas AF, Rajagopal VM, Als TD et al (2019) Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet 51(4):659–674
Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y, Song L, Safi A et al (2018) Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet 50(4):538–548
Pavlides JM, Zhu Z, Gratten J, McRae AF, Wray NR, Yang J (2016) Predicting gene targets from integrative analyses of summary data from GWAS and eQTL studies for 28 human complex traits. Genome Med 8(1):84
Yang Z, Zhou D, Li H, Cai X, Liu W, Wang L, Chang H, Li M et al. (2020) The genome-wide risk alleles for psychiatric disorders at 3p21.1 show convergent effects on mRNA expression, cognitive function, and mushroom dendritic spine. Mol Psychiatry 25 (1):48–66
GTEx Consortium, Laboratory & Data Analysis & Coordinating Center -Analysis Working Group, Statistical Methods groups-Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, Nih/NCI, NIH/NHGRI, NIH/NIMH et al. (2017) Genetic effects on gene expression across human tissues. Nature 550 (7675):204–213
Zhou D, Jiang Y, Zhong X, Cox NJ, Liu C, Gamazon ER (2020) A unified framework for joint-tissue transcriptome-wide association and Mendelian randomization analysis. Nat Genet 52(11):1239–1246
Smeland OB, Frei O, Dale AM, Andreassen OA (2020) The polygenic architecture of schizophrenia - rethinking pathogenesis and nosology. Nat Rev Neurol 16(7):366–379
Hoftman GD, Datta D, Lewis DA (2017) Layer 3 excitatory and inhibitory circuitry in the prefrontal cortex: developmental trajectories and alterations in schizophrenia. Biol Psychiatry 81(10):862–873
Friedman NP, Robbins TW (2021) The role of prefrontal cortex in cognitive control and executive function. Neuropsychopharmacology
Menon V, D'Esposito M (2021) The role of PFC networks in cognitive control and executive function. Neuropsychopharmacology.
Gothelf D, Soreni N, Nachman RP, Tyano S, Hiss Y, Reiner O, Weizman A (2000) Evidence for the involvement of the hippocampus in the pathophysiology of schizophrenia. Eur Neuropsychopharmacol 10(5):389–395
Andreasen NC, Pierson R (2008) The role of the cerebellum in schizophrenia. Biol Psychiatry 64(2):81–88
Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, Won H, van Bakel H et al. (2018) Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362 (6420):eaat8127
Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z et al. (2018) Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362 (6420):eaat7615
Silbereis JC, Pochareddy S, Zhu Y, Li M, Sestan N (2016) The cellular and molecular landscapes of the developing human central nervous system. Neuron 89(2):248–268
Polioudakis D, de la Torre-Ubieta L, Langerman J, Elkins AG, Shi X, Stein JL, Vuong CK, Nichterwitz S et al (2019) A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron 103(5):785-801.e788
Flutre T, Wen X, Pritchard J, Stephens M (2013) A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet 9 (5):e1003486
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88(1):76–82
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL et al (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44(W1):W90-97
Ulgen E, Ozisik O, Sezerman OU (2019) pathfindR: an R package for comprehensive identification of enriched pathways in omics data through active subnetworks. Front Genet 10:858
Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J et al (2016) Schizophrenia risk from complex variation of complement component 4. Nature 530(7589):177–183
Yilmaz M, Yalcin E, Presumey J, Aw E, Ma M, Whelan CW, Stevens B, McCarroll SA et al (2020) Overexpression of schizophrenia susceptibility factor human complement C4A promotes excessive synaptic loss and behavioral changes in mice. Nat Neurosci 24(2):214–224
Wu Y, Li X, Liu J, Luo XJ, Yao YG (2020) SZDB2.0: an updated comprehensive resource for schizophrenia research. Hum Genet 139 (10):1285–1297
Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, Jansen R, de Geus EJ et al (2016) Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48(3):245–252
van Os J, Kapur S (2009) Schizophrenia Lancet 374(9690):635–645
Owen MJ, Sawa A, Mortensen PB (2016) Schizophrenia Lancet 388(10039):86–97
He X, Fuller CK, Song Y, Meng Q, Zhang B, Yang X, Li H (2013) Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet 92(5):667–680
Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG et al (2011) Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478(7370):519–523
de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH (2016) Advancing the understanding of autism disease mechanisms through genetics. Nat Med 22(4):345–361
Gandal MJ, Leppa V, Won H, Parikshak NN, Geschwind DH (2016) The road to precision psychiatry: translating genetics into disease mechanisms. Nat Neurosci 19(11):1397–1407
Amiri A, Coppola G, Scuderi S, Wu F, Roychowdhury T, Liu F, Pochareddy S, Shin Y et al. (2018) Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science 362 (6420):eaat6720
Wortinger LA, Jorgensen KN, Barth C, Nerland S, Smelror RE, Vaskinn A, Ueland T, Andreassen OA et al. (2021) Significant association between intracranial volume and verbal intellectual abilities in patients with schizophrenia and a history of birth asphyxia. Psychol Med:1–10
Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME et al (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48(5):481–487
Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V (2014) Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10 (5):e1004383
Ma L, Semick SA, Chen Q, Li C, Tao R, Price AJ, Shin JH, Jia Y et al (2020) Schizophrenia risk variants influence multiple classes of transcripts of sorting nexin 19 (SNX19). Mol Psychiatry 25(4):831–843
Li M, Jaffe AE, Straub RE, Tao R, Shin JH, Wang Y, Chen Q, Li C et al. (2016) A human-specific AS3MT isoform and BORCS7 are molecular risk factors in the 10q24.32 schizophrenia-associated locus. Nat Med 22 (6):649–656
Cai X, Yang ZH, Li HJ, Xiao X, Li M, Chang H (2021) A human-specific schizophrenia risk tandem repeat affects alternative splicing of a human-unique isoform AS3MTd2d3 and mushroom dendritic spine density. Schizophr Bull 47(1):219–227
Chang H, Cai X, Li HJ, Liu WP, Zhao LJ, Zhang CY, Wang JY, Liu JW et al. (2021) Functional genomics identify a regulatory risk variation rs4420550 in the 16p11.2 schizophrenia-associated locus. Biol Psychiatry 89 (3):246–255
Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C et al (2006) Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet 38(8):910–916
Muller N (2018) Inflammation in Schizophrenia: Pathogenetic Aspects and Therapeutic Considerations. Schizophr Bull 44(5):973–982
Zhang G, Feenstra B, Bacelis J, Liu X, Muglia LM, Juodakis J, Miller DE, Litterman N et al (2017) Genetic Associations with Gestational Duration and Spontaneous Preterm Birth. N Engl J Med 377(12):1156–1167
Weinberger DR (1987) Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry 44(7):660–669
Weinberger DR (2017) The neurodevelopmental origins of schizophrenia in the penumbra of genomic medicine. World Psychiatry 16(3):225–226
Rapoport JL, Addington AM, Frangou S, Psych MR (2005) The neurodevelopmental model of schizophrenia: update 2005. Mol Psychiatry 10(5):434–449
Fatemi SH, Folsom TD (2009) The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr Bull 35(3):528–548
Birnbaum R, Weinberger DR (2017) Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci 18(12):727–740
Zhang CY, Xiao X, Zhang Z, Hu Z, Li M (2021) An alternative splicing hypothesis for neuropathology of schizophrenia: evidence from studies on historical candidate genes and multi-omics data. Mol Psychiatry. https://doi.org/10.1038/s41380-021-01037-w
Walker RL, Ramaswami G, Hartl C, Mancuso N, Gandal MJ, de la Torre-Ubieta L, Pasaniuc B, Stein JL et al (2019) Genetic control of expression and splicing in developing human brain Informs disease mechanisms. Cell 179(3):750-771.e722
Hannon E, Spiers H, Viana J, Pidsley R, Burrage J, Murphy TM, Troakes C, Turecki G et al (2016) Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci 19(1):48–54
Jaffe AE, Gao Y, Deep-Soboslay A, Tao R, Hyde TM, Weinberger DR, Kleinman JE (2016) Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci 19(1):40–47
Bryois J, Garrett ME, Song L, Safi A, Giusti-Rodriguez P, Johnson GD, Shieh AW, Buil A et al (2018) Evaluation of chromatin accessibility in prefrontal cortex of individuals with schizophrenia. Nat Commun 9(1):3121
de la Torre-Ubieta L, Stein JL, Won H, Opland CK, Liang D, Lu D, Geschwind DH (2018) The dynamic landscape of open chromatin during human cortical neurogenesis. Cell 172(1–2):289-304.e218
Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, Clarke D, Gu M et al. (2018) Comprehensive functional genomic resource and integrative model for the human brain. Science 362 (6420):eaat8464
Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ et al (2016) Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538(7626):523–527
Acknowledgements
For CMC data, data were generated as part of the CommonMind Consortium supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffmann-La Roche Ltd and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881, AG02219, AG05138, MH06692, R01MH110921, R01MH109677, R01MH109897, U01MH103392, and contract HHSN271201300031C through IRP NIMH. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories, and the NIMH Human Brain Collection Core. CMC Leadership: Panos Roussos, Joseph Buxbaum, Andrew Chess, Schahram Akbarian, Vahram Haroutunian (Icahn School of Medicine at Mount Sinai), Bernie Devlin, David Lewis (University of Pittsburgh), Raquel Gur, Chang-Gyu Hahn (University of Pennsylvania), Enrico Domenici (University of Trento), Mette A. Peters, Solveig Sieberts (Sage Bionetworks), Thomas Lehner, Stefano Marenco, Barbara K. Lipska (NIMH).
For PsychENCODE data, data were generated as part of the PsychENCODE Consortium, supported by: U01MH103392, U01MH103365, U01MH103346, U01MH103340, U01MH103339, R21MH109956, R21MH105881, R21MH105853, R21MH103877, R21MH102791, R01MH111721, R01MH110928, R01MH110927, R01MH110926, R01MH110921, R01MH110920, R01MH110905, R01MH109715, R01MH109677, R01MH105898, R01MH105898, R01MH094714, P50MH106934, U01MH116488, U01MH116487, U01MH116492, U01MH116489, U01MH116438, U01MH116441, U01MH116442, R01MH114911, R01MH114899, R01MH114901, R01MH117293, R01MH117291, R01MH117292 awarded to: Schahram Akbarian (Icahn School of Medicine at Mount Sinai), Gregory Crawford (Duke University), Stella Dracheva (Icahn School of Medicine at Mount Sinai), Peggy Farnham (University of Southern California), Mark Gerstein (Yale University), Daniel Geschwind (University of California, Los Angeles), Fernando Goes (Johns Hopkins University), Thomas M. Hyde (Lieber Institute for Brain Development), Andrew Jaffe (Lieber Institute for Brain Development), James A. Knowles (University of Southern California), Chunyu Liu (SUNY Upstate Medical University), Dalila Pinto (Icahn School of Medicine at Mount Sinai), Panos Roussos (Icahn School of Medicine at Mount Sinai), Stephan Sanders (University of California, San Francisco), Nenad Sestan (Yale University), Pamela Sklar (Icahn School of Medicine at Mount Sinai), Matthew State (University of California, San Francisco), Patrick Sullivan (University of North Carolina), Flora Vaccarino (Yale University), Daniel Weinberger (Lieber Institute for Brain Development), Sherman Weissman (Yale University), Kevin White (University of Chicago), Jeremy Willsey (University of California, San Francisco), and Peter Zandi (Johns Hopkins University).
The Genotype-Tissue Expression (GTEx) project was supported by the Common Fund of the Office of the Director of the National Institutes of Health and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Funding
This study was supported by the Health Commission of Hubei Province scientific research project (WJ2021M025 to Y.W.) and Health Commission of Wuhan scientific research project (WX20Q02 to Y.W.).
Author information
Authors and Affiliations
Contributions
Y.W. and Y.L. designed the study and interpreted the results. Y.W. and X.L.Y. conducted all the data analysis. Y.W., M.L. and X.X. drafted the manuscript, and all authors contributed to the final version of the paper.
Corresponding authors
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All authors have seen and approved the manuscript and contributed significantly to this work.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, Y., Yu, XL., Xiao, X. et al. Joint-Tissue Integrative Analysis Identified Hundreds of Schizophrenia Risk Genes. Mol Neurobiol 59, 107–116 (2022). https://doi.org/10.1007/s12035-021-02572-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-021-02572-x