Abstract
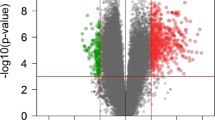
Fingolimod (FTY), a second-line oral drug approved for relapsing remitting Multiple Sclerosis (RRMS) acts in preventing lymphocyte migration outside lymph nodes; moreover, several lines of evidence suggest that it also inhibits myeloid cell activation. In this study, we investigated the transcriptional changes induced by FTY in monocytes in order to better elucidate its mechanism of action. CD14+ monocytes were collected from 24 RRMS patients sampled at baseline and after 6 months of treatment and RNA profiles were obtained through next-generation sequencing. We conducted pathway and sub-paths analysis, followed by centrality analysis of cell-specific interactomes on differentially expressed genes (DEGs). We investigated also the predictive role of baseline monocyte transcription profile in influencing the response to FTY therapy. We observed a marked down-regulation effect (60 down-regulated vs. 0 up-regulated genes). Most of the down-regulated DEGs resulted related with monocyte activation and migration like IL7R, CCR7 and the Wnt signaling mediators LEF1 and TCF7. The involvement of Wnt signaling was also confirmed by subpaths analyses. Furthermore, pathway and network analyses showed an involvement of processes related to immune function and cell migration. Baseline transcriptional profile of the HLA class II gene HLA-DQA1 and HLA-DPA1 were associated with evidence of disease activity after 2 years of treatment. Our data support the evidence that FTY induces major transcriptional changes in monocytes, mainly regarding genes involved in cell trafficking and immune cell activation. The baseline transcriptional levels of genes associated with antigen presenting function were associated with disease activity after 2 years of FTY treatment.


Similar content being viewed by others
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
References
Filippi M, Bar-Or A, Piehl F et al (2018) Multiple sclerosis. Nat Rev Dis Prim 4:43. https://doi.org/10.1038/s41572-018-0041-4
Compston A, Coles A (2008) Multiple sclerosis. Lancet 372:1502–1517. https://doi.org/10.1016/S0140-6736(08)61620-7
Mishra MK, Yong VW (2016) Myeloid cells — targets of medication in multiple sclerosis. Nat Rev Neurol 12:539–551. https://doi.org/10.1038/nrneurol.2016.110
Patel AA, Zhang Y, Fullerton JN et al (2017) The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med 214:1913–1923. https://doi.org/10.1084/jem.20170355
Jakubzick CV, Randolph GJ, Henson PM (2017) Monocyte differentiation and antigen-presenting functions. Nat Rev Immunol 17:349–362. https://doi.org/10.1038/nri.2017.28
Stoeckle C, Tolosa E (2010) Antigen processing and presentation in multiple sclerosis. Results Probl Cell Differ 51:149–172. https://doi.org/10.1007/400_2009_22
Ajami B, Bennett JL, Krieger C et al (2011) Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat Neurosci 14:1142–1149. https://doi.org/10.1038/nn.2887
Mildner A, Mack M, Schmidt H et al (2009) CCR2+Ly-6Chi monocytes are crucial for the effector phase of autoimmunity in the central nervous system. Brain 132:2487–2500. https://doi.org/10.1093/brain/awp144
Chun J, Hartung H-P (2010) Mechanism of action of oral fingolimod (FTY720) in multiple sclerosis. Clin Neuropharmacol 33:91–101. https://doi.org/10.1097/WNF.0b013e3181cbf825
Pitteri M, Magliozzi R, Bajrami A et al (2018) Potential neuroprotective effect of Fingolimod in multiple sclerosis and its association with clinical variables. Expert Opin Pharmacother 19:387–395. https://doi.org/10.1080/14656566.2018.1434143
O’Sullivan SA, O’Sullivan C, Healy LM et al (2018) Sphingosine 1-phosphate receptors regulate TLR4-induced CXCL5 release from astrocytes and microglia. J Neurochem 144:736–747. https://doi.org/10.1111/jnc.14313
Golan M, Mausner-Fainberg K, Ibrahim B et al (2019) Fingolimod increases brain-derived neurotrophic factor level secretion from circulating T cells of patients with multiple sclerosis. CNS Drugs 33:1229–1237. https://doi.org/10.1007/s40263-019-00675-7
Luessi F, Kraus S, Trinschek B et al (2015) FTY720 (fingolimod) treatment tips the balance towards less immunogenic antigen-presenting cells in patients with multiple sclerosis. Mult Scler 21:1811–1822. https://doi.org/10.1177/1352458515574895
Di Dario M, Colombo E, Govi C et al (2015) Myeloid cells as target of fingolimod action in multiple sclerosis. Neurol Neuroimmunol neuroinflammation 2:e157. https://doi.org/10.1212/NXI.0000000000000157
Hughes JE, Srinivasan S, Lynch KR et al (2008) Sphingosine-1-phosphate induces an antiinflammatory phenotype in macrophages. Circ Res 102:950–958. https://doi.org/10.1161/CIRCRESAHA.107.170779
Michaud J, Im D-S, Hla T (2010) Inhibitory role of sphingosine 1-phosphate receptor 2 in macrophage recruitment during inflammation. J Immunol 184:1475–1483. https://doi.org/10.4049/jimmunol.0901586
Polman CH, Reingold SC, Banwell B et al (2011) Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 69:292–302. https://doi.org/10.1002/ana.22366
Jokubaitis VG, Li V, Kalincik T et al (2014) Fingolimod after natalizumab and the risk of short-term relapse. Neurology 82:1204–1211. https://doi.org/10.1212/WNL.0000000000000283
Ottoboni L, Keenan BT, Tamayo P et al (2012) An RNA profile identifies two subsets of multiple sclerosis patients differing in disease activity. Sci Transl Med 4:153ra131. https://doi.org/10.1126/scitranslmed.3004186
Bevan CJ, Cree BAC (2014) Disease activity free status: a new end point for a new era in multiple sclerosis clinical research? JAMA Neurol 71:269–270. https://doi.org/10.1001/jamaneurol.2013.5486
Kappos L, Radue E-W, Chin P et al (2016) Onset of clinical and MRI efficacy occurs early after fingolimod treatment initiation in relapsing multiple sclerosis. J Neurol 263:354–360. https://doi.org/10.1007/s00415-015-7978-y
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527. https://doi.org/10.1038/nbt.3519
Soneson C, Love MI, Robinson MD (2015) Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4:1521. https://doi.org/10.12688/f1000research.7563.2
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Hochberg Y, Benjamini Y (1990) More powerful procedures for multiple significance testing. Stat Med 9:811–818. https://doi.org/10.1002/sim.4780090710
Esposito F, Ferrè L, Clarelli F et al (2018) Effectiveness and baseline factors associated to fingolimod response in a real-world study on multiple sclerosis patients. J Neurol 265:896–905. https://doi.org/10.1007/s00415-018-8791-1
Wang J, Vasaikar S, Shi Z et al (2017) WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. https://doi.org/10.1093/nar/gkx356
Mitrea C, Taghavi Z, Bokanizad B et al (2013) Methods and approaches in the topology-based analysis of biological pathways. Front Physiol 4:278. https://doi.org/10.3389/fphys.2013.00278
Koumakis L, Kanterakis A, Kartsaki E et al (2016) MinePath: mining for phenotype differential sub-paths in molecular pathways. PLoS Comput Biol 12:e1005187. https://doi.org/10.1371/journal.pcbi.1005187
Beisser D, Klau GW, Dandekar T et al (2010) BioNet: an R-Package for the functional analysis of biological networks. Bioinformatics 26:1129–1130. https://doi.org/10.1093/bioinformatics/btq089
Scardoni G, Tosadori G, Faizan M et al (2014) Biological network analysis with CentiScaPe: centralities and experimental dataset integration. F1000Res 3:139. https://doi.org/10.12688/f1000research.4477.2
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Newman AM, Steen CB, Liu CL et al (2019) Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 37:773–782. https://doi.org/10.1038/s41587-019-0114-2
Angerer IC, Hecker M, Koczan D et al (2018) Transcriptome profiling of peripheral blood immune cell populations in multiple sclerosis patients before and during treatment with a sphingosine-1-phosphate receptor modulator. CNS Neurosci Ther 24:193–201. https://doi.org/10.1111/cns.12793
Friess J, Hecker M, Roch L et al (2017) Fingolimod alters the transcriptome profile of circulating CD4+ cells in multiple sclerosis. Sci Rep 7:42087. https://doi.org/10.1038/srep42087
Roch L, Hecker M, Friess J et al (2017) High-resolution expression profiling of peripheral blood CD8+ cells in patients with multiple sclerosis displays fingolimod-induced immune cell redistribution. Mol Neurobiol 54:5511–5525. https://doi.org/10.1007/s12035-016-0075-0
Moreno-Torres I, González-García C, Marconi M et al (2018) Immunophenotype and transcriptome profile of patients with multiple sclerosis treated with fingolimod: setting up a model for prediction of response in a 2-year translational study. Front Immunol 9:1693. https://doi.org/10.3389/fimmu.2018.01693
Ingersoll MA, Spanbroek R, Lottaz C et al (2010) Comparison of gene expression profiles between human and mouse monocyte subsets. Blood 115:e10–e19. https://doi.org/10.1182/blood-2009-07-235028
Randolph GJ, Beaulieu S, Lebecque S et al (1998) Differentiation of monocytes into dendritic cells in a model of transendothelial trafficking. Science 282:480–483. https://doi.org/10.1126/science.282.5388.480
Jakubzick C, Gautier EL, Gibbings SL et al (2013) Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity 39:599–610. https://doi.org/10.1016/j.immuni.2013.08.007
Kim TS, Braciale TJ (2009) Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PLoS ONE 4:e4204. https://doi.org/10.1371/journal.pone.0004204
Zigmond E, Varol C, Farache J et al (2012) Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 37:1076–1090. https://doi.org/10.1016/j.immuni.2012.08.026
Hohl TM, Rivera A, Lipuma L et al (2009) Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe 6:470–481. https://doi.org/10.1016/j.chom.2009.10.007
Sponaas A-M, Freitas do Rosario AP, Voisine C, et al (2009) Migrating monocytes recruited to the spleen play an important role in control of blood stage malaria. Blood 114:5522–5531. https://doi.org/10.1182/blood-2009-04-217489
Nagai T, Devergne O, Mueller TF et al (2003) Timing of IFN-beta exposure during human dendritic cell maturation and naive Th cell stimulation has contrasting effects on Th1 subset generation: a role for IFN-beta-mediated regulation of IL-12 family cytokines and IL-18 in naive Th cell differentiation. J Immunol 171:5233–5243. https://doi.org/10.4049/jimmunol.171.10.5233
Thomas K, Sehr T, Proschmann U et al (2017) Fingolimod additionally acts as immunomodulator focused on the innate immune system beyond its prominent effects on lymphocyte recirculation. J Neuroinflammation 14:41. https://doi.org/10.1186/s12974-017-0817-6
Müller H, Hofer S, Kaneider N et al (2005) The immunomodulator FTY720 interferes with effector functions of human monocyte-derived dendritic cells. Eur J Immunol 35:533–545. https://doi.org/10.1002/eji.200425556
Lan YY, Tokita D, Wang Z et al (2008) Sphingosine 1-phosphate receptor agonism impairs skin dendritic cell migration and homing to secondary lymphoid tissue: association with prolonged allograft survival. Transpl Immunol 20:88–94. https://doi.org/10.1016/j.trim.2008.07.004
Zuvich RL, McCauley JL, Oksenberg JR et al (2010) Genetic variation in the IL7RA/IL7 pathway increases multiple sclerosis susceptibility. Hum Genet 127:525–535. https://doi.org/10.1007/s00439-010-0789-4
Todd JA, Walker NM, Cooper JD et al (2007) Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 39:857–864. https://doi.org/10.1038/ng2068
O’Doherty C, Alloza I, Rooney M, Vandenbroeck K (2009) IL7RA polymorphisms and chronic inflammatory arthropathies. Tissue Antigens 74:429–431. https://doi.org/10.1111/j.1399-0039.2009.01342.x
Heron M, Grutters JC, van Moorsel CHM et al (2009) Variation in IL7R predisposes to sarcoid inflammation. Genes Immun 10:647–653. https://doi.org/10.1038/gene.2009.55
Hoffjan S, Beygo J, Akkad DA et al (2009) Analysis of variation in the IL7RA and IL2RA genes in atopic dermatitis. J Dermatol Sci 55:138–140. https://doi.org/10.1016/j.jdermsci.2009.05.001
Mells GF, Floyd JAB, Morley KI et al (2011) Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet 43:329–332. https://doi.org/10.1038/ng.789
Liu X, Leung S, Wang C et al (2010) Crucial role of interleukin-7 in T helper type 17 survival and expansion in autoimmune disease. Nat Med 16:191–197. https://doi.org/10.1038/nm.2077
Fry TJ, Mackall CL (2002) Interleukin-7: from bench to clinic. Blood 99:3892–3904. https://doi.org/10.1182/blood.v99.11.3892
Standiford TJ, Strieter RM, Allen RM et al (1992) IL-7 up-regulates the expression of IL-8 from resting and stimulated human blood monocytes. J Immunol 149:2035–2039
Alderson MR, Tough TW, Ziegler SF, Grabstein KH (1991) Interleukin 7 induces cytokine secretion and tumoricidal activity by human peripheral blood monocytes. J Exp Med 173:923–930. https://doi.org/10.1084/jem.173.4.923
Chen Z, Kim S, Chamberlain ND et al (2013) The novel role of IL-7 ligation to IL-7 receptor in myeloid cells of rheumatoid arthritis and collagen-induced arthritis. J Immunol 190:5256–5266. https://doi.org/10.4049/jimmunol.1201675
Lorenowicz MJ, van Gils J, de Boer M et al (2006) Epac1-Rap1 signaling regulates monocyte adhesion and chemotaxis. J Leukoc Biol 80:1542–1552. https://doi.org/10.1189/jlb.0506357
Uhlen M, Karlsson MJ, Zhong W et al (2019) A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 366(6472):eaax9198. https://doi.org/10.1126/science.aax9198
Disanto G, Kjetil Sandve G, Ricigliano VAG et al (2014) DNase hypersensitive sites and association with multiple sclerosis. Hum Mol Genet 23:942–948. https://doi.org/10.1093/hmg/ddt489
Mazzola MA, Raheja R, Murugaiyan G et al (2015) Identification of a novel mechanism of action of fingolimod (FTY720) on human effector T cell function through TCF-1 upregulation. J Neuroinflammation 12:245. https://doi.org/10.1186/s12974-015-0460-z
Al-Mossawi H, Yager N, Taylor CA et al (2019) Context-specific regulation of surface and soluble IL7R expression by an autoimmune risk allele. Nat Commun 10:4575. https://doi.org/10.1038/s41467-019-12393-1
Eastman Q, Grosschedl R (1999) Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr Opin Cell Biol 11:233–240. https://doi.org/10.1016/s0955-0674(99)80031-3
Reya T, Clevers H (2005) Wnt signalling in stem cells and cancer. Nature 434:843–850. https://doi.org/10.1038/nature03319
Reya T, O’Riordan M, Okamura R et al (2000) Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity 13:15–24. https://doi.org/10.1016/s1074-7613(00)00004-2
Yu S, Zhou X, Steinke FC et al (2012) The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity 37:813–826. https://doi.org/10.1016/j.immuni.2012.08.009
Xue H-H, Zhao D-M (2012) Regulation of mature T cell responses by the Wnt signaling pathway. Ann N Y Acad Sci 1247:16–33. https://doi.org/10.1111/j.1749-6632.2011.06302.x
Staal FJT, Sen JM (2008) The canonical Wnt signaling pathway plays an important role in lymphopoiesis and hematopoiesis. Eur J Immunol 38:1788–1794. https://doi.org/10.1002/eji.200738118
Tickenbrock L, Schwäble J, Strey A et al (2006) Wnt signaling regulates transendothelial migration of monocytes. J Leukoc Biol 79:1306–1313. https://doi.org/10.1189/jlb.0905539
van de Laar L, van den Bosch A, van der Kooij SW et al (2010) A nonredundant role for canonical NF-κB in human myeloid dendritic cell development and function. J Immunol 185:7252–7261. https://doi.org/10.4049/jimmunol.1000672
Frankenberger M, Pforte A, Sternsdorf T et al (1994) Constitutive nuclear NF-kappa B in cells of the monocyte lineage. Biochem J 304(Pt 1):87–94. https://doi.org/10.1042/bj3040087
Takashiba S, Van Dyke TE, Amar S et al (1999) Differentiation of monocytes to macrophages primes cells for lipopolysaccharide stimulation via accumulation of cytoplasmic nuclear factor kappaB. Infect Immun 67:5573–5578. https://doi.org/10.1128/IAI.67.11.5573-5578.1999
Harhaj EW, Dixit VM (2012) Regulation of NF-κB by deubiquitinases. Immunol Rev 246:107–124. https://doi.org/10.1111/j.1600-065X.2012.01100.x
Ellrichmann G, Thöne J, Lee D-H et al (2012) Constitutive activity of NF-kappa B in myeloid cells drives pathogenicity of monocytes and macrophages during autoimmune neuroinflammation. J Neuroinflammation 9:15. https://doi.org/10.1186/1742-2094-9-15
Steimle A, Kalbacher H, Maurer A et al (2016) A novel approach for reliable detection of cathepsin S activities in mouse antigen presenting cells. J Immunol Methods 432:87–94. https://doi.org/10.1016/j.jim.2016.02.015
Haves-Zburof D, Paperna T, Gour-Lavie A et al (2011) Cathepsins and their endogenous inhibitors cystatins: expression and modulation in multiple sclerosis. J Cell Mol Med 15:2421–2429. https://doi.org/10.1111/j.1582-4934.2010.01229.x
Grossman I, Avidan N, Singer C et al (2007) Pharmacogenetics of glatiramer acetate therapy for multiple sclerosis reveals drug-response markers. Pharmacogenet Genomics 17:657–666. https://doi.org/10.1097/FPC.0b013e3281299169
Cunningham S, Graham C, Hutchinson M et al (2005) Pharmacogenomics of responsiveness to interferon IFN-beta treatment in multiple sclerosis: a genetic screen of 100 type I interferon-inducible genes. Clin Pharmacol Ther 78:635–646. https://doi.org/10.1016/j.clpt.2005.08.018
Karagkouni A, Alevizos M, Theoharides TC (2013) Effect of stress on brain inflammation and multiple sclerosis. Autoimmun Rev 12:947–953. https://doi.org/10.1016/j.autrev.2013.02.006
van Langelaar J, Rijvers L, Smolders J, van Luijn MM (2020) B and T cells driving multiple sclerosis: identity, mechanisms and potential triggers. Front Immunol 11:760. https://doi.org/10.3389/fimmu.2020.00760
Kurashima Y, Kunisawa J, Higuchi M et al (2007) Sphingosine 1-phosphate-mediated trafficking of pathogenic Th2 and mast cells for the control of food allergy. J Immunol 179:1577–1585. https://doi.org/10.4049/jimmunol.179.3.1577
Kleinjan A, van Nimwegen M, Leman K et al (2013) Topical treatment targeting sphingosine-1-phosphate and sphingosine lyase abrogates experimental allergic rhinitis in a murine model. Allergy 68:204–212. https://doi.org/10.1111/all.12082
Acknowledgements
We wish to thank all the patients, family members and nurses of the San Raffaele Hospital MS centre that participated in the study.
Funding
The study was supported by a grant from Fondazione Italiana Sclerosi Multipla (grant number FISM2013/R/13).
Author information
Authors and Affiliations
Contributions
GS contributed to the clinical characterization of studied patients, performed pathway analyses, contributed to the interpretation of the results and wrote the manuscript. FC contributed to study design, carried out bioinformatic analyses and wrote the manuscript. EM carried out cell separation and the RNASeq experiments and contributed to the interpretation of the results. LF enrolled the patients involved in the study and contributed to the interpretation of the results. LO, MS, SS contributed to the interpretation of the results. LM, VM, GC, MF and FMB contributed to patient enrollment and discussion of article content. PP carried out the bioinformatic and differential expression analyses and contributed the discussion of results. FE designed the experiments, contributed to the discussion of results and wrote the manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
The local independent Ethical Committee approved the study protocol and all patients signed the informed consent before enrolling in the study.
Consent for Publication
Not applicable.
Competing Interests
G. Sferruzza, F. Clarelli, E. Mascia, L. Ferrè, L. Ottoboni, M. Sorosina, S. Santoro, P. Provero report no competing financial interest.
L. Moiola received honoraria for speaking or partecipating to advisory board from TEVA, Novartis, Biogen, Sanofi e Roche.
V. Martinelli has received honoraria for consulting services or speaking activity from Biogen, Merck, Novartis, TEVA, Almirall, and Genzyme.
G. Comi has received G. Comi has received personal compensation for consulting and speaking activities from Novartis, Teva, Sanofi Genzyme, Merck, Biogen, Roche, Almirall, Celgene, Forward Pharma, Medday and Excemed.
F. Martinelli Boneschi has received compensation for activities with Teva Neuroscience, Biogen Idec, Merck Serono as speaker and/or advisor.
M Filippi is Editor-in-Chief of the Journal of Neurology; received compensation for consulting services and/or speaking activities from Biogen Idec, Merck-Serono, Novartis, Teva Pharmaceutical Industries; and receives research support from Biogen Idec, Merck-Serono, Novartis, Teva Pharmaceutical Industries, Roche, Italian Ministry of Health, Fondazione Italiana Sclerosi Multipla, and ARiSLA (Fondazione Italiana di Ricerca per la SLA).
F. Esposito received honoraria from Novartis and received research support from the Italian Ministry of Health, Fondazione Italiana Sclerosi Multipla and European Union (Horizon 2020 Research and Innovation programme).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
G. Sferruzza and F. Clarelli contributed equally to the study.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sferruzza, G., Clarelli, F., Mascia, E. et al. Transcriptomic Analysis of Peripheral Monocytes upon Fingolimod Treatment in Relapsing Remitting Multiple Sclerosis Patients. Mol Neurobiol 58, 4816–4827 (2021). https://doi.org/10.1007/s12035-021-02465-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-021-02465-z