Abstract
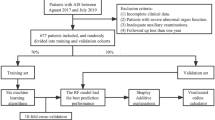
Although delayed cerebral ischemia (DCI) is a well-known complication after subarachnoid hemorrhage (SAH), there are no reliable biomarkers to predict DCI development. Matricellular proteins (MCPs) have been reported relevant to DCI and expected to become biomarkers. As machine learning (ML) enables the classification of various input data and the result prediction, the aim of this study was to construct early prediction models of DCI development with clinical variables and MCPs using ML analyses. Early-stage clinical data of 95 SAH patients in a prospective cohort were analyzed and applied to a ML algorithm, random forest, to construct three prediction models: (1) a model with only clinical variables on admission, (2) a model with only plasma levels of MCP (periostin, osteopontin, and galectin-3) at post-onset days 1–3, and (3) a model with both clinical variables on admission and MCP values at days 1–3. The prediction accuracy of the development of DCI, angiographic vasospasm, or cerebral infarction and the importance of each feature were computed. The prediction accuracy of DCI development was 93.9% in model 1, 87.2% in model 2, and 95.1% in model 3, but that of angiographic vasospasm or cerebral infarction was lower. The three most important features in model 3 for DCI were periostin, osteopontin, and galectin-3, followed by aneurysm location. All of the early-stage prediction models of DCI development constructed by ML worked with high accuracy and sensitivity. One-time early-stage measurement of plasma MCPs served for reliable prediction of DCI development, suggesting their potential utility as biomarkers.



Similar content being viewed by others
References
Vergouwen MDI, Vermeulen M, van Gijn J, Rinkel GJE, Wijdicks EF, Muizelaar JP, Mendelow AD, Juvela S et al (2010) Definition of delayed cerebral ischemia after aneurysmal subarachnoid hemorrhage as an outcome event in clinical trials and observational studies: proposal of a multidisciplinary research group. Stroke 41:2391–2395. https://doi.org/10.1161/STROKEAHA.110.589275
Foreman PM, Chua MH, Harrigan MR, Fisher WS, Tubbs RS, Shoja MM, Griessenauer CJ (2017) External validation of the practical risk chart for the prediction of delayed cerebral ischemia following aneurysmal subarachnoid hemorrhage. J Neurosurg 126:1530–1536. https://doi.org/10.3171/2016.1.JNS152554
de Rooij NK, Greving JP, Rinkel GJE, Frijns CJM (2013) Early prediction of delayed cerebral ischemia after subarachnoid hemorrhage: development and validation of a practical risk chart. Stroke 44:1288–1294. https://doi.org/10.1161/STROKEAHA.113.001125
Lee H, Perry JJ, English SW, Alkherayf F, Joseph J, Nobile S, Zhou LL, Lesiuk H et al (2018) Clinical prediction of delayed cerebral ischemia in aneurysmal subarachnoid hemorrhage. J Neurosurg. https://doi.org/10.3171/2018.1.JNS172715 [published Online First: 8 June 2018]
de Oliveira Manoel AL, Jaja BN, Germans MR, Yan H, Qian W, Kouzmina E, Marotta TR, Turkel-Parrella D et al (2015) The VASOGRADE: a simple grading scale for prediction of delayed cerebral ischemia after subarachnoid hemorrhage. Stroke 46:1826–1831. https://doi.org/10.1161/STROKEAHA.115.008728
Crobeddu E, Mittal MK, Dupont S, Wijdicks EFM, Lanzino G, Rabinstein AA (2012) Predicting the lack of development of delayed cerebral ischemia after aneurysmal subarachnoid hemorrhage. Stroke 43:697–701. https://doi.org/10.1161/STROKEAHA.111.638403
Suzuki H, Nishikawa H, Kawakita F (2018) Matricellular proteins as possible biomarkers for early brain injury after aneurysmal subarachnoid hemorrhage. Neural Regen Res 13:1175–1178. https://doi.org/10.4103/1673-5374.235022
Murphy-Ullrich JE, Sage EH (2014) Revisiting the matricellular concept. Matrix Biol 37:1–14. https://doi.org/10.1016/j.matbio.2014.07.005
Nakatsuka Y, Shiba M, Nishikawa H et al (2018) Acute-phase plasma osteopontin as an independent predictor for poor outcome after aneurysmal subarachnoid hemorrhage. Mol Neurobiol 55:6841–6849. https://doi.org/10.1007/s12035-018-0893-3
Nishikawa H, Nakatsuka Y, Shiba M et al (2018) Increased plasma galectin-3 preceding the development of delayed cerebral infarction and eventual poor outcome in non-severe aneurysmal subarachnoid hemorrhage. Transl Stroke Res 9:110–119. https://doi.org/10.1007/s12975-017-0564-0
Nishikawa H, Suzuki H (2017) Implications of periostin in the development of subarachnoid hemorrhage-induced brain injuries. Neural Regen Res 12:1982–1984. https://doi.org/10.4103/1673-5374.221150
Nishikawa H, Suzuki H (2018) Possible role of inflammation and galectin-3 in brain injury after subarachnoid hemorrhage. Brain Sci 8. https://doi.org/10.3390/brainsci8020030
Brusko GD, Kolcun JPG, Wang MY (2018) Machine-learning models: the future of predictive analytics in neurosurgery. Neurosurgery 83:E3–E4. https://doi.org/10.1093/neuros/nyy166
Muhlestein WE, Akagi DS, Davies JM, Chambless LB (2018) Predicting inpatient length of stay after brain tumor surgery: developing machine learning ensembles to improve predictive performance. Neurosurgery. https://doi.org/10.1093/neuros/nyy343 [published Online First: 3 August 2018]
Senders JT, Arnaout O, Karhade AV, Dasenbrock HH, Gormley WB, Broekman ML, Smith TR (2018) Natural and artificial intelligence in neurosurgery: a systematic review. Neurosurgery 83:181–192. https://doi.org/10.1093/neuros/nyx384
Müller AC, Guido S (2017) Introduction to machine learning with Python: a guide for data scientists. 1st ed. O’Reilly, Sebastopol
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357. https://doi.org/10.1613/jair.953
Random forest classifier. scikit-learn. https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html. Accessed 8 Feb 2019
Louppe G (2015) Understanding random forests: from theory to practice. arXiv. https://arxiv.org/abs/1407.7502. Accessed 8 Feb 2019
Ensemble methods. scikit-learn. https://scikit-learn.org/stable/modules/ensemble.html. Accessed 8 Feb 2019
Suzuki H, Hasegawa Y, Kanamaru K, Zhang JH (2010) Mechanisms of osteopontin-induced stabilization of blood-brain barrier disruption after subarachnoid hemorrhage in rats. Stroke 41:1783–1790. https://doi.org/10.1161/STROKEAHA.110.586537
Suzuki H (2015) What is early brain injury? Transl Stroke Res 6:1–3. https://doi.org/10.1007/s12975-014-0380-8
Suzuki H, Ayer R, Sugawara T, Chen W, Sozen T, Hasegawa Y, Kanamaru K, Zhang JH (2010) Protective effects of recombinant osteopontin on early brain injury after subarachnoid hemorrhage in rats. Crit Care Med 38:612–618. https://doi.org/10.1097/CCM.0b013e3181c027ae
Suzuki H, Hasegawa Y, Chen W, Kanamaru K, Zhang JH (2010) Recombinant osteopontin in cerebral vasospasm after subarachnoid hemorrhage. Ann Neurol 68:650–660. https://doi.org/10.1002/ana.22102
Topkoru BC, Altay O, Duris K, Krafft PR, Yan J, Zhang JH (2013) Nasal administration of recombinant osteopontin attenuates early brain injury after subarachnoid hemorrhage. Stroke 44:3189–3194. https://doi.org/10.1161/STROKEAHA.113.001574
Liu L, Kawakita F, Fujimoto M, Nakano F, Imanaka-Yoshida K, Yoshida T, Suzuki H (2017) Role of periostin in early brain injury after subarachnoid hemorrhage in mice. Stroke 48:1108–1111. https://doi.org/10.1161/STROKEAHA.117.016629
Nishikawa H, Liu L, Nakano F, Kawakita F, Kanamaru H, Nakatsuka Y, Okada T, Suzuki H (2018) Modified citrus pectin prevents blood-brain barrier disruption in mouse subarachnoid hemorrhage by inhibiting galectin-3. Stroke 49:2743–2751. https://doi.org/10.1161/STROKEAHA.118.021757
Kanamaru H, Kawakita F, Nakano F et al (2019) Plasma periostin and delayed cerebral ischemia after aneurysmal subarachnoid hemorrhage. Neurotherapeutics. https://doi.org/10.1007/s13311-018-00707-y
Yoshimoto Y, Kwak S (1995) Age-related multifactorial causes of neurological deterioration after early surgery for aneurysmal subarachnoid hemorrhage. J Neurosurg 83:984–988. https://doi.org/10.3171/jns.1995.83.6.0984
Lanzino G, Kassell NF, Germanson TP, Kongable GL, Truskowski LL, Torner JC, Jane JA (1996) Age and outcome after aneurysmal subarachnoid hemorrhage: why do older patients fare worse? J Neurosurg 85:410–418. https://doi.org/10.3171/jns.1996.85.3.0410
Macdonald RL, Kassell NF, Mayer S, Ruefenacht D, Schmiedek P, Weidauer S, Frey A, Roux S et al (2008) Clazosentan to overcome neurological ischemia and infarction occurring after subarachnoid hemorrhage (CONSCIOUS-1): randomized, double-blind, placebo-controlled phase 2 dose-finding trial. Stroke 39:3015–3021. https://doi.org/10.1161/STROKEAHA.108.519942
Rosalind Lai PM, Du R (2015) Role of genetic polymorphisms in predicting delayed cerebral ischemia and radiographic vasospasm after aneurysmal subarachnoid hemorrhage: a meta-analysis. World Neurosurg 84:933–941.e2. https://doi.org/10.1016/j.wneu.2015.05.070
Suzuki H, Shiba M, Nakatsuka Y, Nakano F, Nishikawa H (2017) Higher cerebrospinal fluid pH may contribute to the development of delayed cerebral ischemia after aneurysmal subarachnoid hemorrhage. Transl Stroke Res 8:165–173. https://doi.org/10.1007/s12975-016-0500-8
Cui Z, Gong G (2018) The effect of machine learning regression algorithms and sample size on individualized behavioral prediction with functional connectivity features. Neuroimage 178:622–637. https://doi.org/10.1016/j.neuroimage.2018.06.001
Chan MTH, Wong JYY, Leung AKT et al (2019) Plasma and CSF miRNA dysregulations in subarachnoid hemorrhage reveal clinical courses and underlying pathways. J Clin Neurosci 62:155–161. https://doi.org/10.1016/j.jocn.2018.11.038
Jabbarli R, Pierscianek D, Darkwah Oppong M, Sato T, Dammann P, Wrede KH, Kaier K, Köhrmann M et al (2018) Laboratory biomarkers of delayed cerebral ischemia after subarachnoid hemorrhage: a systematic review. Neurosurg Rev. https://doi.org/10.1007/s10143-018-1037-y [published Online First: 10 October 2018]
Alghamdi M, Al-Mallah M, Keteyian S et al (2017) Predicting diabetes mellitus using SMOTE and ensemble machine learning approach: The Henry Ford ExercIse Testing (FIT) roject. PLoS One 12:e0179805. https://doi.org/10.1371/journal.pone.0179805
Sui Y, Wei Y, Zhao D (2015) Computer-aided lung nodule recognition by SVM classifier based on combination of random undersampling and SMOTE. Comput Math Methods Med 368674:1–13. https://doi.org/10.1155/2015/368674
Acknowledgements
We thank Dr. Nobuhisa Kashiwagi (the Institute of Statistical Mathematics, Tokyo, Japan) for review of statistical analyses.
Funding
This work was supported by a grant-in-aid for Scientific Research from Japan Society for the Promotion of Science to Dr. HS (grant number 17K10825).
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(PDF 684 kb)
Rights and permissions
About this article
Cite this article
Tanioka, S., Ishida, F., Nakano, F. et al. Machine Learning Analysis of Matricellular Proteins and Clinical Variables for Early Prediction of Delayed Cerebral Ischemia After Aneurysmal Subarachnoid Hemorrhage. Mol Neurobiol 56, 7128–7135 (2019). https://doi.org/10.1007/s12035-019-1601-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-019-1601-7