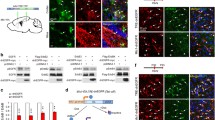
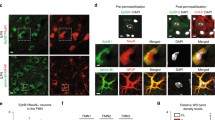
Abstract
In this study, we found that Sp1 was highly expressed in astrocytes, implying that Sp1 might be important for the function of astrocytes. Sp1/GFAP-Cre-ERT2 conditional knockout mice were constructed to study the role of Sp1 in astrocytes. Knockout of Sp1 in astrocytes altered astrocytic morphology and decreased GFAP expression in the cortex and hippocampus but did not affect cell viability. Loss of Sp1 in astrocytes decreased the number of neurons in the cortex and hippocampus. Conditioned medium from primary astrocytes with Sp1 knockout disrupted neuronal dendritic outgrowth and synapse formation, resulting in abnormal learning, memory, and motor behavior. Sp1 knockout in astrocytes altered gene expression, including decreasing the expression of Toll-like receptor 2 and Cfb and increasing the expression of C1q and C4Bp, thereby affecting neurite outgrowth and synapse formation, resulting in disordered neuron function. Studying these gene regulations might be beneficial to understanding neuronal development and brain injury prevention.









Similar content being viewed by others
References
Li L, Davie JR (2010) The role of Sp1 and Sp3 in normal and cancer cell biology. Ann Anat 192(5):275–283. https://doi.org/10.1016/j.aanat.S0940-9602(10)00116-0
Wang SA, Chuang JY, Yeh SH, Wang YT, Liu YW, Chang WC, Hung JJ (2009) Heat shock protein 90 is important for Sp1 stability during mitosis. J Mol Biol 387(5):1106–1119. https://doi.org/10.1016/j.jmb.2009.02.040
Lagger G, Doetzlhofer A, Schuettengruber B, Haidweger E, Simboeck E, Tischler J, Chiocca S, Suske G et al (2003) The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol Cell Biol 23(8):2669–2679
Opitz OG, Rustgi AK (2000) Interaction between Sp1 and cell cycle regulatory proteins is important in transactivation of a differentiation-related gene. Cancer Res 60(11):2825–2830
Wei C, Zhang W, Zhou Q, Zhao C, Du Y, Yan Q, Li Z, Miao J (2016) Mithramycin A alleviates cognitive deficits and reduces neuropathology in a transgenic mouse model of Alzheimer’s disease. Neurochem Res 41(8):1924–1938. https://doi.org/10.1007/s11064-016-1903-310.1007/s11064-016-1903-3
Wang J, Song W (2016) Regulation of LRRK2 promoter activity and gene expression by Sp1. Mol Brain 9:33. https://doi.org/10.1186/s13041-016-0215-510.1186/s13041-016-0215-5
Citron BA, Saykally JN, Cao C, Dennis JS, Runfeldt M, Arendash GW (2015) Transcription factor Sp1 inhibition, memory, and cytokines in a mouse model of Alzheimer’s disease. Am J Neurodegener Dis 4(2):40–48
Chuang JY, Kao TJ, Lin SH, Wu AC, Lee PT, Su TP, Yeh SH, Lee YC et al (2017) Specificity protein 1-zinc finger protein 179 pathway is involved in the attenuation of oxidative stress following brain injury. Redox Biol 11:135–143. https://doi.org/10.1016/j.redox.2016.11.012
Miras-Portugal MT, Gomez-Villafuertes R, Gualix J, Diaz-Hernandez JI, Artalejo AR, Ortega F, Delicado EG, Perez-Sen R (2016) Nucleotides in neuroregeneration and neuroprotection. Neuropharmacology 104:243–254. https://doi.org/10.1016/j.neuropharm.2015.09.002S0028-3908(15)30094-0
Simard JM, Chen M, Tarasov KV, Bhatta S, Ivanova S, Melnitchenko L, Tsymbalyuk N, West GA et al (2006) Newly expressed SUR1-regulated NC(Ca-ATP) channel mediates cerebral edema after ischemic stroke. Nat Med 12(4):433–440. https://doi.org/10.1038/nm1390
Yeh SH, Yang WB, Gean PW, Hsu CY, Tseng JT, Su TP, Chang WC, Hung JJ (2011) Translational and transcriptional control of Sp1 against ischaemia through a hydrogen peroxide-activated internal ribosomal entry site pathway. Nucleic Acids Res 39(13):5412–5423. https://doi.org/10.1093/nar/gkr161
Eroglu C, Barres BA (2010) Regulation of synaptic connectivity by glia. Nature 468(7321):223–231. https://doi.org/10.1038/nature09612nature09612
Chung WS, Allen NJ, Eroglu C (2015) Astrocytes control synapse formation, function, and elimination. Cold Spring Harb Perspect Biol 7(9):a020370. https://doi.org/10.1101/cshperspect.a020370
Malarkey EB, Parpura V (2008) Mechanisms of glutamate release from astrocytes. Neurochem Int 52(1–2):142–154. https://doi.org/10.1016/j.neuint.2007.06.005
Haydon PG, Carmignoto G (2006) Astrocyte control of synaptic transmission and neurovascular coupling. Physiol Rev 86(3):1009–1031. https://doi.org/10.1152/physrev.00049.2005
Allen NJ, Eroglu C (2017) Cell biology of astrocyte-synapse interactions. Neuron 96(3):697–708. https://doi.org/10.1016/j.neuron.2017.09.056
Li B, Chen P, Qu J, Shi L, Zhuang W, Fu J, Li J, Zhang X et al (2014) Activation of LTBP3 gene by a long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem cells from multiple myeloma. J Biol Chem 289(42):29365–29375. https://doi.org/10.1074/jbc.M114.572693M114.572693
Yang WB, Chen PH, Hsu TS, Fu TF, Su WC, Liaw H, Chang WC, Hung JJ (2014) Sp1-mediated microRNA-182 expression regulates lung cancer progression. Oncotarget 5(3):740–753. https://doi.org/10.18632/oncotarget.1608
Kruger I, Vollmer M, Simmons DG, Elsasser HP, Philipsen S, Suske G (2007) Sp1/Sp3 compound heterozygous mice are not viable: impaired erythropoiesis and severe placental defects. Dev Dyn 236(8):2235–2244. https://doi.org/10.1002/dvdy.21222
Marin M, Karis A, Visser P, Grosveld F, Philipsen S (1997) Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 89(4):619–628
Michinaga S, Ishida A, Takeuchi R, Koyama Y (2013) Endothelin-1 stimulates cyclin D1 expression in rat cultured astrocytes via activation of Sp1. Neurochem Int 63(1):25–34. https://doi.org/10.1016/j.neuint.2013.04.004
Jin M, Ande A, Kumar A, Kumar S (2013) Regulation of cytochrome P450 2e1 expression by ethanol: role of oxidative stress-mediated pkc/jnk/sp1 pathway. Cell Death Dis 4:e554. https://doi.org/10.1038/cddis.2013.78cddis201378
Loeffler S, Fayard B, Weis J, Weissenberger J (2005) Interleukin-6 induces transcriptional activation of vascular endothelial growth factor (VEGF) in astrocytes in vivo and regulates VEGF promoter activity in glioblastoma cells via direct interaction between STAT3 and Sp1. Int J Cancer 115(2):202–213. https://doi.org/10.1002/ijc.20871
Choi SS, Lee HJ, Lim I, Satoh J, Kim SU (2014) Human astrocytes: secretome profiles of cytokines and chemokines. PLoS One 9(4):e92325. https://doi.org/10.1371/journal.pone.0092325PONE-D-13-48616
Jakel S, Dimou L (2017) Glial cells and their function in the adult brain: a journey through the history of their ablation. Front Cell Neurosci 11:24. https://doi.org/10.3389/fncel.2017.00024
Yamakawa H, Cheng J, Penney J, Gao F, Rueda R, Wang J, Yamakawa S, Kritskiy O et al (2017) The transcription factor Sp3 cooperates with HDAC2 to regulate synaptic function and plasticity in neurons. Cell Rep 20(6):1319–1334. https://doi.org/10.1016/j.celrep.2017.07.044
Ishimaru N, Tabuchi A, Hara D, Hayashi H, Sugimoto T, Yasuhara M, Shiota J, Tsuda M (2007) Regulation of neurotrophin-3 gene transcription by Sp3 and Sp4 in neurons. J Neurochem 100(2):520–531. https://doi.org/10.1111/j.1471-4159.2006.04216.x
Mao XR, Moerman-Herzog AM, Chen Y, Barger SW (2009) Unique aspects of transcriptional regulation in neurons—nuances in NFkappaB and Sp1-related factors. J Neuroinflammation 6:16. https://doi.org/10.1186/1742-2094-6-16
Lian H, Yang L, Cole A, Sun L, Chiang AC, Fowler SW, Shim DJ, Rodriguez-Rivera J et al (2015) NFkappaB-activated astroglial release of complement C3 compromises neuronal morphology and function associated with Alzheimer’s disease. Neuron 85(1):101–115. https://doi.org/10.1016/j.neuron.2014.11.018
Shastri A, Bonifati DM, Kishore U (2013) Innate immunity and neuroinflammation. Mediators Inflamm 2013:342931. https://doi.org/10.1155/2013/342931
Okun E, Griffioen KJ, Lathia JD, Tang SC, Mattson MP, Arumugam TV (2009) Toll-like receptors in neurodegeneration. Brain Res Rev 59(2):278–292. https://doi.org/10.1016/j.brainresrev.2008.09.001S0165-0173(08)00103-3
Gesuete R, Kohama SG, Stenzel-Poore MP (2014) Toll-like receptors and ischemic brain injury. J Neuropathol Exp Neurol 73(5):378–386. https://doi.org/10.1097/NEN.0000000000000068
Jacob A, Alexander JJ (2014) Complement and blood-brain barrier integrity. Mol Immunol 61(2):149–152. https://doi.org/10.1016/j.molimm.2014.06.039S0161-5890(14)00170-9
Rutkowski MJ, Sughrue ME, Kane AJ, Ahn BJ, Fang S, Parsa AT (2010) The complement cascade as a mediator of tissue growth and regeneration. Inflamm Res 59(11):897–905. https://doi.org/10.1007/s00011-010-0220-6
Boulanger LM (2009) Immune proteins in brain development and synaptic plasticity. Neuron 64(1):93–109. https://doi.org/10.1016/j.neuron.2009.09.001S0896-6273(09)00678-3
Veerhuis R, Nielsen HM, Tenner AJ (2011) Complement in the brain. Mol Immunol 48(14):1592–1603. https://doi.org/10.1016/j.molimm.2011.04.003S0161-5890(11)00120-9
Fraser DA, Pisalyaput K, Tenner AJ (2010) C1q enhances microglial clearance of apoptotic neurons and neuronal blebs, and modulates subsequent inflammatory cytokine production. J Neurochem 112(3):733–743. https://doi.org/10.1111/j.1471-4159.2009.06494.xJNC6494
Tokudome K, Okumura T, Shimizu S, Mashimo T, Takizawa A, Serikawa T, Terada R, Ishihara S et al (2016) Synaptic vesicle glycoprotein 2A (SV2A) regulates kindling epileptogenesis via GABAergic neurotransmission. Sci Rep 6:27420. https://doi.org/10.1038/srep27420srep27420
Wood IC, Garriga M, Palmer CL, Pepitoni S, Buckley NJ (1999) Neuronal expression of the rat M1 muscarinic acetylcholine receptor gene is regulated by elements in the first exon. Biochem J 340(Pt 2):475–483
Lou XY, Ma JZ, Payne TJ, Beuten J, Crew KM, Li MD (2006) Gene-based analysis suggests association of the nicotinic acetylcholine receptor beta1 subunit (CHRNB1) and M1 muscarinic acetylcholine receptor (CHRM1) with vulnerability for nicotine dependence. Hum Genet 120(3):381–389. https://doi.org/10.1007/s00439-006-0229-7
Ma DQ, Whitehead PL, Menold MM, Martin ER, Ashley-Koch AE, Mei H, Ritchie MD, Delong GR et al (2005) Identification of significant association and gene-gene interaction of GABA receptor subunit genes in autism. Am J Hum Genet 77(3):377–388. https://doi.org/10.1086/433195
Greene ND, Copp AJ (2014) Neural tube defects. Annu Rev Neurosci 37:221–242. https://doi.org/10.1146/annurev-neuro-062012-170354
Kalashnikova E, Lorca RA, Kaur I, Barisone GA, Li B, Ishimaru T, Trimmer JS, Mohapatra DP et al (2010) SynDIG1: an activity-regulated, AMPA- receptor-interacting transmembrane protein that regulates excitatory synapse development. Neuron 65(1):80–93. https://doi.org/10.1016/j.neuron.2009.12.021S0896-6273(09)01011-3
Maloyan A, Sanbe A, Osinska H, Westfall M, Robinson D, Imahashi K, Murphy E, Robbins J (2005) Mitochondrial dysfunction and apoptosis underlie the pathogenic process in alpha-B-crystallin desmin-related cardiomyopathy. Circulation 112(22):3451–3461. https://doi.org/10.1161/CIRCULATIONAHA.105.572552
Garcia-Huerta P, Diaz-Hernandez M, Delicado EG, Pimentel-Santillana M, Miras-Portugal MT, Gomez-Villafuertes R (2012) The specificity protein factor Sp1 mediates transcriptional regulation of P2X7 receptors in the nervous system. J Biol Chem 287(53):44628–44644. https://doi.org/10.1074/jbc.M112.390971M112.390971
Paonessa F, Latifi S, Scarongella H, Cesca F, Benfenati F (2013) Specificity protein 1 (Sp1)-dependent activation of the synapsin I gene (SYN1) is modulated by RE1-silencing transcription factor (REST) and 5′-cytosine-phosphoguanine (CpG) methylation. J Biol Chem 288(5):3227–3239. https://doi.org/10.1074/jbc.M112.399782M112.399782
Boutillier S, Lannes B, Buee L, Delacourte A, Rouaux C, Mohr M, Bellocq JP, Sellal F et al (2007) Sp3 and sp4 transcription factor levels are increased in brains of patients with Alzheimer’s disease. Neurodegener Dis 4(6):413–423. https://doi.org/10.1159/000107701
Saia G, Lalonde J, Sun X, Ramos B, Gill G (2014) Phosphorylation of the transcription factor Sp4 is reduced by NMDA receptor signaling. J Neurochem 129(4):743–752. https://doi.org/10.1111/jnc.12657
Ramos B, Gaudilliere B, Bonni A, Gill G (2007) Transcription factor Sp4 regulates dendritic patterning during cerebellar maturation. Proc Natl Acad Sci U S A 104(23):9882–9887. https://doi.org/10.1073/pnas.0701946104
Aoyama T, Okamoto T, Fukiage K, Otsuka S, Furu M, Ito K, Jin Y, Ueda M et al (2010) Histone modifiers, YY1 and p300, regulate the expression of cartilage-specific gene, chondromodulin-I, in mesenchymal stem cells. J Biol Chem 285(39):29842–29850. https://doi.org/10.1074/jbc.M110.116319M110.116319
Yao YL, Yang WM, Seto E (2001) Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol 21(17):5979–5991
Han JW, Ahn SH, Kim YK, Bae GU, Yoon JW, Hong S, Lee HY, Lee YW et al (2001) Activation of p21(WAF1/Cip1) transcription through Sp1 sites by histone deacetylase inhibitor apicidin: involvement of protein kinase C. J Biol Chem 276(45):42084–42090. https://doi.org/10.1074/jbc.M106688200M106688200
Billon N, Carlisi D, Datto MB, van Grunsven LA, Watt A, Wang XF, Rudkin BB (1999) Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene 18(18):2872–2882. https://doi.org/10.1038/sj.onc.1202712
Acknowledgments
We are grateful for the support of clinical specimens from the Human Biobank, Research Center of Clinical Medicine, and National Cheng Kung University Hospital.
Funding
This work was supported by the grants (106-2320-B-006-065-MY3, 106-2320-B-006-020-MY3, and 104-2923-B-038-002-MY3) obtained from the Ministry of Science and Technology, Taiwan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary figure 1
Sp1 expression level in Sp1 knockout mice. Wild type, Sp1f/+ and Sp1f/f mice were treated with tamoxifen and then sacrificed. The Sp1 level was studied by Western blot with anti-Sp1 antibodies. The Sp1 level was quantified and statistical assay by t-test, *<0.05, **<0.01. (JPG 467 kb)
Supplementary figure 2
Sp1-mediated pathway and molecular function in astrocyte. Sp1 was knockout from astrocyte by tamoxifen treatment, and then the RNA was isolated for performing the cDNA array. The gene expression involved in pathways (A) and molecular function (B) was analysis by Gene Ontology analysis. (JPG 760 kb)
Supplementary figure 3
Sp1 in astrocyte regulates the pathways and various biological processes in neurons. Conditional medium was collected from wild type astrocyte or Sp1 knockout astrocyte, and then treated the neurons. The RNA was isolated for performing the cDNA array. The gene expression involved in pathways (A) and biological processes (B) was analysis by Gene Ontology analysis. (JPG 1038 kb)
Supplementary figure 4
Sp1 in astrocyte regulates the group of proteins in neurons. The proteomic clusters was analyzed in neuron treated with conditional medium from astrocytes with or without Sp1 knockout. (JPG 1042 kb)
Rights and permissions
About this article
Cite this article
Hung, CY., Hsu, TI., Chuang, JY. et al. Sp1 in Astrocyte Is Important for Neurite Outgrowth and Synaptogenesis. Mol Neurobiol 57, 261–277 (2020). https://doi.org/10.1007/s12035-019-01694-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-019-01694-7