Abstract
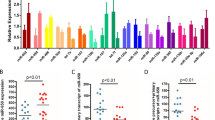
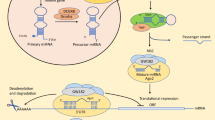
Cyclin-dependent kinase 5 regulatory subunit 1 (CDK5R1) encodes p35, the main activatory subunit of cyclin-dependent kinase 5 (CDK5). The p35/CDK5 active complex plays a fundamental role in brain development and functioning, but its deregulated activity has also been implicated in various neurodegenerative disorders, including Alzheimer’s disease (AD). CDK5R1 displays a large and highly evolutionarily conserved 3′-untranslated region (3′-UTR), a fact that has suggested a role for this region in the post-transcriptional control of CDK5R1 expression. Our group has recently demonstrated that two miRNAs, miR-103 and miR-107, regulate CDK5R1 expression and affect the levels of p35. MiR-103 and miR-107 belong to the miR-15/107 family, a group of evolutionarily conserved miRNAs highly expressed in human cerebral cortex. In this work, we tested the hypothesis that other members of this group of miRNAs, in addition to miR-103 and miR-107, were able to modulate CDK5R1 expression. We provide evidence that several miRNAs belonging to the miR-15/107 family regulate p35 levels. BACE1 expression levels were also found to be modulated by different members of this family. Furthermore, overexpression of these miRNAs led to reduced APP phosphorylation levels at the CDK5-specific Thr668 residue. We also show that miR-15/107 miRNAs display reduced expression levels in hippocampus and temporal cortex, but not in cerebellum, of AD brains. Moreover, increased CDK5R1 mRNA levels were observed in AD hippocampus tissues. Our results suggest that the downregulation of the miR-15/107 family might have a role in the pathogenesis of AD by increasing the levels of CDK5R1/p35 and consequently enhancing CDK5 activity.







Similar content being viewed by others
References
Su SC, Tsai LH (2011) Cyclin-dependent kinases in brain development and disease. Annu Rev Cell Dev Biol 27:465–491
Ohshima T, Ward JM, Huh CG, Longenecker G, Veeranna, Pant HC, Brady RO, Martin LJ et al (1996) Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc Natl Acad Sci U S A 93:11173–11178
Chae T, Kwon YT, Bronson R, Dikkes P, Li E, Tsai LH (1997) Mice lacking p35, a neuronal specific activator of Cdk5, display cortical lamination defects, seizures, and adult lethality. Neuron 18:29–42
McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM (1984) Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34:939–944
Qiu C, Kivipelto M, von Strauss E (2009) Epidemiology of Alzheimer’s disease: occurrence, determinants, and strategies toward intervention. Dialogues Clin Neurosci 11:111–128
Chouraki V, Seshadri S (2014) Genetics of Alzheimer’s disease. Adv Genet 87:245–294
Jiang T, Chang RC, Rosenmann H, Yu JT (2015) Advances in Alzheimer’s disease: from bench to bedside. Biomed Res Int 2015:202676
Patrick GN, Zukerberg L, Nikolic M, de la Monte S, Dikkes P, Tsai LH (1999) Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature 402:615–622
Kusakawa G, Saito T, Onuki R, Ishiguro K, Kishimoto T, Hisanaga S (2000) Calpain-dependent proteolytic cleavage of the p35 cyclin-dependent kinase 5 activator to p25. J Biol Chem 275:17166–17172
Ahlijanian MK, Barrezueta NX, Williams RD, Jakowski A, Kowsz KP, McCarthy S, Coskran T, Carlo A et al (2000) Hyperphosphorylated tau and neurofilament and cytoskeletal disruptions in mice overexpressing human p25, an activator of cdk5. Proc Natl Acad Sci U S A 97:2910–2915
Cruz JC, Tseng HC, Goldman JA, Shih H, Tsai LH (2003) Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron 40:471–483
Piedrahita D, Hernández I, López-Tobón A, Fedorov D, Obara B, Manjunath BS, Boudreau RL, Davidson B et al (2010) Silencing of CDK5 reduces neurofibrillary tangles in transgenic alzheimer’s mice. J Neurosci 30:13966–13976
Iijima K, Ando K, Takeda S, Satoh Y, Seki T, Itohara S, Greengard P, Kirino Y et al (2000) Neuron-specific phosphorylation of Alzheimer’s beta-amyloid precursor protein by cyclin-dependent kinase 5. J Neurochem 75:1085–1091
Shukla V, Skuntz S, Pant HC (2012) Deregulated Cdk5 activity is involved in inducing Alzheimer’s disease. Arch Med Res 43:655–662
Moncini S, Bevilacqua A, Venturin M, Fallini C, Ratti A, Nicolin A, Riva P (2007) The 3′ untranslated region of human Cyclin-Dependent Kinase 5 Regulatory subunit 1 contains regulatory elements affecting transcript stability. BMC Mol Biol 8:111
Mignone F, Gissi C, Liuni S, Pesole G (2002) Untranslated regions of mRNAs. Genome Biol 3:REVIEWS0004
Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z (2003) The microRNA world: small is mighty. Trends Biochem Sci 28:534–540
Kosik KS, Krichevsky AM (2005) The elegance of the MicroRNAs: a neuronal perspective. Neuron 47:779–782
Nelson PT, Wang WX, Rajeev BW (2008) MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol 18:130–138
Satoh J (2010) MicroRNAs and their therapeutic potential for human diseases: aberrant microRNA expression in Alzheimer’s disease brains. J Pharmacol Sci 114:269–275
Salta E, De Strooper B (2012) Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol 11:189–200
Tan L, Yu JT, Hu N, Tan L (2013) Non-coding RNAs in Alzheimer’s disease. Mol Neurobiol 47:382–393
Moncini S, Salvi A, Zuccotti P, Viero G, Quattrone A, Barlati S, De Petro G, Venturin M et al (2011) The role of miR-103 and miR-107 in regulation of CDK5R1 expression and in cellular migration. PLoS One 6, e20038
Finnerty JR, Wang WX, Hébert SS, Wilfred BR, Mao G, Nelson PT (2010) The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles inhuman diseases. J Mol Biol 402:491–509
Wang WX, Danaher RJ, Miller CS, Berger JR, Nubia VG, Wilfred BS, Neltner JH, Norris CM et al (2014) Expression of miR-15/107 family microRNAs in human tissues and cultured rat brain cells. Genomics Proteomics Bioinformatics 12:19–30
Hébert SS, Horré K, Nicolaï L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A et al (2008) Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A 105:6415–6420
Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT (2008) The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci 28:1213–1223
Hébert SS, Papadopoulou AS, Smith P, Galas MC, Planel E, Silahtaroglu AN, Sergeant N, Buée L et al (2010) Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet 19:3959–3969
Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ (2010) Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS One 5, e8898
Liu W, Liu C, Zhu J, Shu P, Yin B, Gong Y, Qiang B, Yuan J et al (2012) MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiol Aging 33:522–534
Zhu HC, Wang LM, Wang M, Song B, Tan S, Teng JF, Duan DX (2012) MicroRNA-195 downregulates Alzheimer’s disease amyloid-β production by targeting BACE1. Brain Res Bull 88:596–601
Ai J, Sun LH, Che H, Zhang R, Zhang TZ, Wu WC, Su XL, Chen X et al (2013) MicroRNA-195 protects against dementia induced by chronic brain hypoperfusion via its anti-amyloidogenic effect in rats. J Neurosci 33:3989–4001
Penna I, Vella S, Gigoni A, Russo C, Cancedda R, Pagano A (2011) Selection of candidate housekeeping genes for normalization in human postmortem brain samples. Int J Mol Sci 12:5461–5470
Linnertz C, Saucier L, Ge D, Cronin KD, Burke JR, Browndyke JN, Hulette CM, Welsh-Bohmer KA et al (2009) Genetic regulation of alpha-synuclein mRNA expression in various human brain tissues. PLoS One 4, e7480
Michalova E, Vojtesek B, Hrstka R (2013) Impaired pre-mRNA processing and altered architecture of 3′ untranslated regions contribute to the development of human disorders. Int J Mol Sci 14:15681–15694
Pillai RS, Bhattacharyya SN, Filipowicz W (2007) Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 17:118–126
Yao J, Hennessey T, Flynt A, Lai E, Beal MF, Lin MT (2010) MicroRNA-related cofilin abnormality in Alzheimer’s disease. PLoS One 5, e15546
Sun LH, Ban T, Liu CD, Chen QX, Wang X, Yan ML, Hu XL, Su XL et al (2015) Activation of Cdk5/p25 and tau phosphorylation following chronic brain hypoperfusion in rats involves microRNA-195 downregulation. J Neurochem 134:1139–1151
Parsi S, Smith PY, Goupil C, Dorval V, Hébert SS (2015) Preclinical evaluation of miR-15/107 family members as multifactorial drug targets for Alzheimer’s Disease. Mol Ther Nucleic Acids 4, e256
Wang WX, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, Stromberg AJ, Huang Q et al (2010) miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am J Pathol 177:334–345
Wang WX, Kyprianou N, Wang X, Nelson PT (2010) Dysregulation of the mitogen granulin in human cancer through the miR-15/107 microRNA gene group. Cancer Res 70:9137–9142
Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J et al (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442:916–919
Zuccotti P, Colombrita C, Moncini S, Barbieri A, Lunghi M, Gelfi C, De Palma S, Nicolin A et al (2014) hnRNPA2/B1 and nELAV proteins bind to a specific U-rich element in CDK5R1 3′-UTR and oppositely regulate its expression. Biochim Biophys Acta 1839:506–516
Mizukami K, Ishikawa M, Iwakiri M, Ikonomovic MD, Dekosky ST, Kamma H, Asada T (2005) Immunohistochemical study of the hnRNP A2 and B1 in the hippocampal formations of brains with Alzheimer’s disease. Neurosci Lett 386:111–115
Amadio M, Pascale A, Wang J, Ho L, Quattrone A, Gandy S, Haroutunian V, Racchi M et al (2009) nELAV proteins alteration in Alzheimer’s disease brain: a novel putative target for amyloid-beta reverberating on AbetaPP processing. J Alzheimers Dis 16:409–419
Acknowledgments
The authors would like to thank the MRC London Neurodegenerative Diseases Brain Bank, the Newcastle Brain Tissue Resource, and the South West Dementia Brain Bank (SWDBB) for providing brain tissue for this study. The SWDBB is supported by BRACE (Bristol Research into Alzheimer’s and Care of the Elderly), Brains for Dementia Research and the Medical Research Council. This work was supported by Ministero dell’Istruzione, dell’Università e della Ricerca (RBFR0895DC_02, FIRB (Basic Research Investment Fund) 2008 Grant to MV and RBFR0895DC_01, FIRB (Basic Research Investment Fund) 2008 Grant to MAD).
Conflict of Interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Table S1
Oligonucleotide pairs designed to generate wild-type and mutated miR-sensor constructs (XLSX 10 kb)
Fig. S1
Validation of miRNA sensors. a Luciferase activity of miRNA sensor constructs in SK-N-BE cells normalized on pmirGLO-empty vector. b Luciferase activity of representative miRNA sensors with a sequence perfectly complementary to the endogenous miRNA (wt) or with a 4-base mutation in the seed sequence (mut). c Luciferase activity of sensor constructs cotransfected with 5 nM of the specific miRNA antisense LNA molecule. *p < 0.05, **p < 0.01, Student’s t test (GIF 1 kb)
Fig. S2
Levels of miR-15/107 miRNA expression in AD and control brain tissues. The expression levels of miR-15/107 miRNAs in three different brain areas (temporal cortex, hippocampus, and cerebellum) of AD patients (n = 12) and controls (n = 12) are represented as relative expression to the small nucleolar RNA RNU6B (GIF 352 bytes)
Fig. S3
Levels of CDK5R1 and BACE1 expression in AD and control brain tissues. The expression levels of CDK5R1 and BACE1 genes in three different brain areas (temporal cortex, hippocampus, and cerebellum) of AD patients (n = 12) and controls (n = 12) are represented as relative expression to the SYP gene. *p < 0.05, Student’s t test (GIF 725 bytes)
Rights and permissions
About this article
Cite this article
Moncini, S., Lunghi, M., Valmadre, A. et al. The miR-15/107 Family of microRNA Genes Regulates CDK5R1/p35 with Implications for Alzheimer’s Disease Pathogenesis. Mol Neurobiol 54, 4329–4342 (2017). https://doi.org/10.1007/s12035-016-0002-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-016-0002-4