Abstract
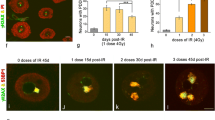
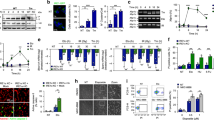
Neurons are highly vulnerable to genotoxic agents. To restore genome integrity upon DNA lesions, neurons trigger a DNA damage response (DDR) that requires chromatin modifications and transcriptional silencing at DNA damage sites. To study the reorganization of the active RNA polymerase II (Pol II), which transcribes all mRNA-encoding genes, and the participation of the ubiquitin-proteasome system (UPS) in the neuronal DDR, we have used rat sensory ganglion neurons exposed to X-rays (4 Gy) ionizing radiation (IR). In control neurons, Pol II appears concentrated in numerous chromatin microfoci identified as transcription factories by the incorporation of 5′-fluorouridine into nascent RNA. Upon IR treatment, numerous IR-induced foci (IRIF), which were immunoreactive for γH2AX and 53BP1, were observed as early as 30 min post-IR; their number progressively reduced at 3 h, 1 day, and 3 days post-IR. The formation of IRIF was associated with a decrease in Pol II levels by both immunofluorescence and Western blotting. Treatment with the proteasome inhibitor bortezomib strongly increased Pol II levels in both control and irradiated neurons, suggesting that proteasome plays a proteolytic role by clearing stalled Pol II complexes at DNA damage sites, as a prelude to DNA repair. Neuronal IRIF recruited ubiquitylated proteins, including ubiquitylated histone H2A (Ub-H2A), and the catalytic proteasome 20S. Ub-H2A has been associated with transcriptional silencing at DNA damage sites. On the other hand, the participation of UPS in neuronal DDR may be essential for the ubiquitylation of Pol II and other proteasome substrates of the DNA repair machinery and their subsequent proteasome-mediated degradation.





Similar content being viewed by others
References
Ptashne M, Gann A (1997) Transcriptional activation by recruitment. Nature 386:569–77
Saunders A, Core LJ, Lis JT (2006) Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol 7:557–67
Jonkers I, Lis JT (2015) Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16:167–77
Hocine S, Singer RH, Grünwald D (2010) RNA processing and export. Cold Spring Harb Perspect Biol 2, a000752
Komarnitsky P, Cho EJ, Buratowski S (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14:2452–60
Warren SL, Landolfi AS, Curtis C, Morrow JS (1992) Cytostellin: a novel, highly conserved protein that undergoes continuous redistribution during the cell cycle. J Cell Sci 103:381–8
Guillot PV, Xie SQ, Hollinshead M, Pombo A (2004) Fixation-induced redistribution of hyperphosphorylated RNA polymerase II in the nucleus of human cells. Exp Cell Res 295:460–8
Ratner JN, Balasubramanian B, Corden J, Warren SL, Bregman DB (1998) Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J Biol Chem 273:5184–9
McKay BC, Chen F, Clarke ST, Wiggin HE, Harley LM, Ljungman M (2001) UV light-induced degradation of RNA polymerase II is dependent on the Cockayne’s syndrome A and B proteins but not p53 or MLH1. Mutat Res 485:93–105
Ljungman M, Lane DP (2004) Transcription - guarding the genome by sensing DNA damage. Nat Rev Cancer 4:727–37
Somesh BP, Reid J, Liu WF, Sogaard TM, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2005) Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121:913–23
Svejstrup JQ (2003) Rescue of arrested RNA polymerase II complexes. J Cell Sci 116:447–51
Pankotai T, Bonhomme C, Chen D, Soutoglou E (2012) DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol 19:276–82
Donahue BA, Yin S, Taylor JS, Reines D, Hanawalt PC (1994) Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci U S A 91:8502–6
Wilson MD, Harreman M, Taschner M, Reid J, Walker J, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2013) Proteasome-mediated processing of Def1, a critical step in the cellular response to transcription stress. Cell 154:983–95
Wilson MD, Harreman M, Svejstrup JQ (2013) Ubiquitylation and degradation of elongating RNA polymerase II: the last resort. Biochim Biophys Acta 1829:151–7
Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131:887–900
Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G et al (2004) Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell 16:1027–34
Englander EW (2013) DNA damage response in peripheral nervous system: coping with cancer therapy-induced DNA lesions. DNA Repair 12:685–90
Ferrer I, Serrano T, Alcantara S, Tortosa A, Graus F (1993) X-ray-induced cell death in the developing hippocampal complex involves neurons and requires protein synthesis. J Neuropathol Exp Neurol 52:370–8
Casafont I, Palanca A, Lafarga V, Berciano MT, Lafarga M (2011) Effect of ionizing radiation in sensory ganglion neurons: organization and dynamics of nuclear compartments of DNA damage/repair and their relationship with transcription and cell cycle. Acta Neuropathol 122:481–493
Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Ann Rev Biochem 79:181–211
Jeppesen DK, Bohr VA, Stevnsner T (2011) DNA repair deficiency in neurodegeneration. Prog Neurobiol 94:166–200
Rass U, Ahel I, West SC (2007) Defective DNA repair and neurodegeneration disease. Cell 130:991–1004
McKinnon PJ (2013) Maintaining genome stability in the nervous system. Nat Neurosci 16:1523–9
Baltanas F, Casafont I, Lafarga V, Weruaga E, Alonso JR, Berciano MT, Lafarga M (2011) Purkinje cell degeneration in pcd mice reveals large scale chromatin reorganization and gene silencing linked to defective DNA repair. J Biol Chem 286:28287–28302
Pan L, Penney J, Tsai LH (2014) Chromatin regulation of DNA damage repair and genome integrity in the central nervous system. J Mol Biol 426:3376–88
Hetman M, Vashishta A, Rempala G (2010) Neurotoxic mechanisms of DNA damage: focus on transcriptional inhibition. J Neurochem 114:1537–1549
Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, Cech TR (2012) FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev 26:2690–5
Pena E, Berciano MT, Fernandez R, Ojeda JL, Lafarga M (2001) Neuronal body size correlates with the number of nucleoli and Cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons. J Comp Neurol 430:250–63
Mengual E, Arizti P, Rodrigo J, Giménez-Amaya JM, Castaño JG (1996) Immunohistochemical distribution and electron microscopic subcellular localization of the proteasome in the rat CNS. J Neurosci 16:6331–41
Lafarga M, Berciano MT, Pena E, Mayo I, Castaño JG, Bohmann D, Rodrigues JP, Tavanez JP et al (2002) Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol Biol Cell 13:2771–82
Casafont I, Navascués J, Pena E, Lafarga M, Berciano MT (2006) Nuclear organization and dynamics of transcription sites in rat sensory ganglia neurons detected by incorporation of 5′-fluorouridine into nascent RNA. Neuroscience 140:453–62
Spector DL, Lamond AI (2011) Nuclear speckles. Cold Spring Harb Perspect Biol 3, a000646
Iborra FJ, Pombo A, Jackson DA, Cook PR (1996) Active RNA polymerases are localized within discrete transcription “factories’ in human nuclei. J Cell Sci 109:1427–36
Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA et al (2004) Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36:1065–71
Rieder D, Trajanoski Z, McNally JG (2012) Transcription factories. Front Genet 3:221
Raska I, Shaw PJ, Cmarko D (2006) New insights into nucleolar architecture and activity. Int Rev Cytol 255:177–235
Palanca A, Casafont I, Berciano MT, Lafarga M (2014) Reactive nucleolar and Cajal body responses to proteasome inhibition in sensory ganglion neurons. Biochim Biophys Acta 1842:848–59
Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A (2004) H2AX: the histone guardian of genome. DNA Repair 3:959–967
Noon AT, Goodarzi AA (2011) 53BP1-mediated DNA double strand break repair: insert bad pun here. DNA Repair 10:1071–1076
Callen E, di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F et al (2013) 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153:1266–1280
Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA (2010) ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141:970–81
Canals-Hamann AZ, das Neves RP, Reittie JE, Iñiguez C, Soneji S, Enver T, Buckle VJ, Iborra FJ (2013) A biophysical model for transcription factories. BMC Biophys 6:2
Chakalova L, Fraser P (2010) Organization of transcription. Cold Spring Harb Perspect Biol 2, a000729
Bickmore WA, van Steensel B (2013) Genome architecture: domain organization of interphase chromosomes. Cell 152:1270–84
Rieder D, Ploner C, Krogsdam AM, Stocker G, Fischer M, Scheideler M, Dani C, Amri EZ et al (2014) Co-expressed genes prepositioned in spatial neighborhoods stochastically associate with SC35 speckles and RNA polymerase II factories. Cell Mol Life Sci 71:1741–59
Zuleger N, Robson MI, Schirmer EC (2011) The nuclear envelope as a chromatin organizer. Nucleus 2:339–49
Wilczynski GM (2014) Significance of higher-order chromatin architecture for neuronal function and dysfunction. Neuropharmacology 80:28–33
Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang JY, Daigle KR, Fenn JD et al (2011) Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat Neurosci 14:848–56
Gillette TG, Gonzalez F, Delahodde A, Johnston SA, Kodadek T (2004) Physical and functional association of RNA polymerase II and the proteasome. Proc Natl Acad Sci U S A 101:5904–9
Tornaletti S, Hanawalt PC (1999) Effect of DNA lesions on transcription elongation. Biochimie 81:139–46
Lu H, Flores O, Weinmann R, Reinberg D (1991) The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci U S A 88:10004–8
Misteli T, Soutoglou E (2009) The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 10:243–54
Palanca A, Casafont I, Berciano MT, Lafarga M (2014) Proteasome inhibition induces DNA damage and reorganizes nuclear architecture and protein synthesis machinery in sensory ganglion neurons. Cell Mol Life Sci 71:1961–75
Lukas J, Lukas C, Bartek J (2011) More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 13:1161–9
Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S et al (2009) RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136:435–446
Motegi A, Murakawa Y, Takeda S (2009) The vital link between the ubiquitin-proteasome pathway and DNA repair: impact on cancer therapy. Cancer Lett 283:1–9
Jacquemont C, Taniguchi T (2007) Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Res 67:7395–405
Takeshita T, Wu W, Koike A, Fukuda M, Ohta T (2009) Perturbation of DNA repair pathways by proteasome inhibitors corresponds to enhanced chemosensitivity of cells to DNA damage-inducing agents. Cancer Chemother Pharmacol 64:1039–46
Acknowledgments
This work was supported by the following grants: “Dirección General de Investigación” of Spain (BFU2011-23983; BFU2014-54754-P) and “Centro de Investigación Biomédica en Red sobre Enfermedades Neurodegenerativas (CIBERNED; CB06/05/0037)” from Spain.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Research Involving Animals
All procedures were approved by the Bioethical Committee of the University of Cantabria and were carried out according to the directives of the Council of the European Communities and current Spanish legislation.
Additional information
Iñigo Casafont and Ana Palanca contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
(DOCX 834 kb)
Rights and permissions
About this article
Cite this article
Casafont, I., Palanca, A., Lafarga, V. et al. Dynamic Behavior of the RNA Polymerase II and the Ubiquitin Proteasome System During the Neuronal DNA Damage Response to Ionizing Radiation. Mol Neurobiol 53, 6799–6808 (2016). https://doi.org/10.1007/s12035-015-9565-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-015-9565-8