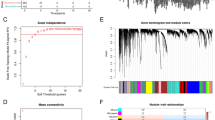
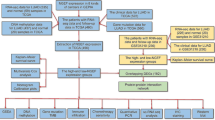
Abstract
Chemoresistance is a key obstacle in the long-term survival of patients with locally and advanced lung adenocarcinoma (LUAD). This study used bioinformatic analysis to reveal the chemoresistance of gene-neutrophil extracellular traps (NETs) associated with LUAD. RNA sequencing data and LUAD expression patterns were obtained from the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases, respectively. The GeneCards database was used to identify NETosis-related genes (NRGs). To identify hub genes with significant and consistent expression, differential analysis was performed using the TCGA-LUAD and GEO datasets. LUAD subtypes were determined based on these hub genes, followed by prognostic analysis. Immunological scoring and infiltration analysis were conducted using NETosis scores (N-scores) derived from the TCGA-LUAD dataset. A clinical prognostic model was established and analyzed, and its clinical applications explored. Twenty-two hub genes were identified, and consensus clustering was used to identify two subgroups based on their expression levels. The Kaplan–Meier (KM) curves demonstrated statistically significant differences in prognosis between the two LUAD subtypes. Based on the median score, patients were further divided into high and low N-score groups, and KM curves showed that the N-scores were more precise at predicting the prognosis of patients with LUAD for overall survival (OS). Immunological infiltration analysis revealed significant differences in the abundances of 10 immune cell infiltrates between the high and low N-score groups. Risk scores indicated significant differences in prognosis between the two extreme score groups. The risk scores for the prognostic model also indicated significant differences between the two groups. The results provide new insights into NETosis-related differentially expressed genes (NRDEGs) associated with chemotherapy resistance in patients with LUAD. The established prognostic model is promising and could help with clinical applications to evaluate patient survival and therapeutic efficiency.









Similar content being viewed by others
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Abbreviations
- LUAD:
-
Lung adenocarcinoma
- NETs:
-
Gene-neutrophil extracellular traps
- TCGA:
-
The Cancer Genome Atlas
- GEO:
-
Gene expression omnibus
- NRGs:
-
NETosis-related genes
- Nscores:
-
NETosis scores
- KM:
-
Kaplan–Meier
- OS:
-
Overall survival
- NRDEGs:
-
NETosis-related differentially expressed genes
- FPKM:
-
Fragments per kilobase per million
- TMB:
-
Tumor mutation burden
- MSI:
-
Microsatellite instability
- SNPs:
-
Single nucleotide polymorphisms
- CNV:
-
Copy number variation
- PPI:
-
Protein–protein interaction
- GO:
-
Gene Ontology
- BP:
-
Biological processes
- MF:
-
Molecular functions
- CC:
-
Cellular components
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- GSVA:
-
Gene set variation analysis
- ssGSEA:
-
Single-sample gene-set enrichment analysis
- TME:
-
Tumor immunological microenvironment
- INS:
-
Number of insertions
- CDF:
-
Cumulative distribution function
- PCA:
-
Principal component analysis
- ROC:
-
Receiver operating characteristic curve
References
Bray, F., Ferlay, J., Soerjomataram, I., et al. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68(6), 394–424.
Li, Y., Yan, B., & He, S. (2023). Advances and challenges in the treatment of lung cancer. Biomedicine & pharmacotherapy Biomedecine & pharmacotherapie, 169, 115891.
Raman, L., Van der Linden, M., Van der Eecken, K., et al. (2020). Shallow whole-genome sequencing of plasma cell-free DNA accurately differentiates small from non-small cell lung carcinoma. Genome Medicine, 12(1), 35.
Miller, M., & Hanna, N. (2021). Advances in systemic therapy for non-small cell lung cancer. BMJ (Clinical Research ed.), 375, n2363.
Lau, M. T., Ghazanfar, S., Parkin, A., et al. (2020). Systematic functional identification of cancer multi-drug resistance genes. Genome Biology, 21(1), 27.
de Bont, C. M., Boelens, W. C., & Pruijn, G. J. M. (2019). NETosis, complement, and coagulation: A triangular relationship. Cellular & Molecular Immunology, 16(1), 19–27.
Masucci, M. T., Minopoli, M., Del Vecchio, S., et al. (2020). The emerging role of neutrophil extracellular traps (NETs) in tumor progression and metastasis. Frontiers in Immunology, 11, 1749.
Cristinziano, L., Modestino, L., Antonelli, A., et al. (2022). Neutrophil extracellular traps in cancer. Seminars in Cancer Biology, 79, 91–104.
Mousset, A., Lecorgne, E., Bourget, I., et al. (2023). Neutrophil extracellular traps formed during chemotherapy confer treatment resistance via TGF-β activation. Cancer Cell, 41(4), 757-775.e10.
Saw, P. E., Chen, J., & Song, E. (2023). ChemoNETosis: A road to tumor therapeutic resistance. Cancer Cell, 41(4), 655–657.
Colaprico, A., Silva, T. C., Olsen, C., et al. (2016). TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Research, 44(8), e71.
Goldman, M. J., Craft, B., Hastie, M., et al. (2020). Visualizing and interpreting cancer genomics data via the Xena platform. Nature Biotechnology, 38(6), 675–678.
Mayakonda, A., Lin, D. C., Assenov, Y., et al. (2018). Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Research, 28(11), 1747–1756.
Mermel, C. H., Schumacher, S. E., Hill, B., et al. (2011). GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biology, 12(4), 41.
Zhang, W., Gong, W., Ai, H., et al. (2014). Gene expression analysis of lung adenocarcinoma and matched adjacent non-tumor lung tissue. Tumori, 100(3), 338–345.
Wei, T. Y., Juan, C. C., Hisa, J. Y., et al. (2012). Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Science, 103(9), 1640–1650.
Barrett, T., Troup, D. B., Wilhite, S. E., et al. (2007). NCBI GEO: Mining tens of millions of expression profiles–database and tools update. Nucleic Acids Research, 35, D760–D765.
Davis, S., & Meltzer, P. S. (2007). GEOquery: A bridge between the gene expression omnibus (GEO) and bioconductor. Bioinformatics (Oxford, England), 23(14), 1846–1847.
Stelzer, G., Rosen, N., Plaschkes, I., et al. (2016). The genecards suite: From gene data mining to disease genome sequence analyses. Current Protocols in Bioinformatics, 54, 1–33.
Love, M. I., Huber, W., & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12), 550.
Szklarczyk, D., Gable, A. L., Lyon, D., et al. (2019). STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research, 47(D1), D607–D613.
Shannon, P., Markiel, A., Ozier, O., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11), 2498–2504.
Franz, M., Rodriguez, H., Lopes, C., et al. (2018). GeneMANIA update 2018. Nucleic Acids Research, 46(W1), W60–W64.
Yu, G. (2020). Gene ontology semantic similarity analysis using GOSemSim. Methods in Molecular Biology, 2117, 207–215.
Kanehisa, M., & Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 28(1), 27–30.
Brière, G., Darbo, É., Thébault, P., et al. (2021). Consensus clustering applied to multi-omics disease subtyping. BMC Bioinformatics, 22(1), 361.
Wilkerson, M. D., & Hayes, D. N. (2010). ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics (Oxford, England), 26(12), 1572–1573.
Hänzelmann, S., Castelo, R., & Guinney, J. (2013). GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics, 14, 7.
Yoshihara, K., Shahmoradgoli, M., Martínez, E., et al. (2013). Inferring tumour purity and stromal and immune cell admixture from expression data. Nature Communications, 4, 2612.
Bindea, G., Mlecnik, B., Tosolini, M., et al. (2013). Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity, 39(4), 782–795.
Newman, A. M., Liu, C. L., Green, M. R., et al. (2015). Robust enumeration of cell subsets from tissue expression profiles. Nature Methods, 12(5), 453–457.
Park, S. Y. (2018). Nomogram: An analogue tool to deliver digital knowledge. The Journal of Thoracic and Cardiovascular Surgery, 155(4), 1793.
Perkins, N. J., Weck, J., Mumford, S. L., et al. (2019). Combining biomarker calibration data to reduce measurement error. Epidemiology, 30(Suppl 2), S3–S9.
Van Calster, B., Wynants, L., Verbeek, J. F. M., et al. (2018). Reporting and interpreting decision curve analysis: A guide for investigators. European Urology, 74(6), 796–804.
Tataranni, T., & Piccoli, C. (2019). Dichloroacetate (DCA) and cancer: An overview towards clinical applications. Oxidative Medicine and Cellular Longevity, 2019, 8201079.
Seguin, L., Durandy, M., & Feral, C. C. (2022). Lung adenocarcinoma tumor origin: A guide for personalized medicine. Cancers, 14(7), 1759.
Zeng, X., Wang, H., He, D., et al. (2020). LIMD1 increases the sensitivity of lung adenocarcinoma cells to cisplatin via the GADD45α/p38 MAPK signaling pathway. Frontiers in Oncology, 10, 969.
Demers, M., Wong, S. L., Martinod, K., et al. (2016). Priming of neutrophils toward NETosis promotes tumor growth. Oncoimmunology, 5(5), e1134073.
Hakkim, A., Fuchs, T. A., Martinez, N. E., et al. (2011). Activation of the Raf-MEK-ERK pathway is required for neutrophil extracellular trap formation. Nature Chemical Biology, 7(2), 75–77.
Shahzad, M. H., Feng, L., Su, X., et al. (2022). Neutrophil extracellular traps in cancer therapy resistance. Cancers, 14(5), 1359.
Ham, I. H., Oh, H. J., Jin, H., et al. (2019). Targeting interleukin-6 as a strategy to overcome stroma-induced resistance to chemotherapy in gastric cancer. Molecular Cancer, 18(1), 68.
Zhai, J., Shen, J., Xie, G., et al. (2019). Cancer-associated fibroblasts-derived IL-8 mediates resistance to cisplatin in human gastric cancer. Cancer Letters, 454, 37–43.
Lee, H. G., Wheeler, M. A., & Quintana, F. J. (2022). Function and therapeutic value of astrocytes in neurological diseases Nature Reviews. Drug discovery, 21(5), 339–358.
Yang, Q., Xu, J., Gu, J., et al. (2022). Extracellular vesicles in cancer drug resistance: Roles, mechanisms, and implications. Advanced Science, 9(34), e2201609.
Zhu, H., Chen, K., Chen, Y., et al. (2022). RNA-binding protein ZCCHC4 promotes human cancer chemoresistance by disrupting DNA-damage-induced apoptosis. Signal Transduction and Targeted Therapy, 7(1), 240.
Low, H. B., Wong, Z. L., Wu, B., et al. (2021). DUSP16 promotes cancer chemoresistance through regulation of mitochondria-mediated cell death. Nature Communications, 12(1), 2284.
Takeuchi, S., Baghdadi, M., Tsuchikawa, T., et al. (2015). Chemotherapy-derived inflammatory responses accelerate the formation of immunosuppressive myeloid cells in the tissue microenvironment of human pancreatic cancer. Cancer Research, 75(13), 2629–2640.
Wesolowski, R., Duggan, M. C., Stiff, A., et al. (2017). Circulating myeloid-derived suppressor cells increase in patients undergoing neo-adjuvant chemotherapy for breast cancer. Cancer Immunology, Immunotherapy : CII, 66(11), 1437–1447.
Galluzzi, L., Humeau, J., Buqué, A., et al. (2020). Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nature Reviews. Clinical Oncology, 17(12), 725–741.
Trivieri, N., Pracella, R., Cariglia, M. G., et al. (2020). BRAFV600E mutation impinges on gut microbial markers defining novel biomarkers for serrated colorectal cancer effective therapies. Journal of Experimental & Clinical Cancer Research : CR, 39(1), 285.
Demaria, O., Cornen, S., Daëron, M., et al. (2019). Harnessing innate immunity in cancer therapy. Nature, 574(7776), 45–56.
Xia, L., Oyang, L., Lin, J., et al. (2021). The cancer metabolic reprogramming and immune response. Molecular Cancer, 20(1), 28.
Palmeri, M., Mehnert, J., Silk, A. W., et al. (2022). Real-world application of tumor mutational burden-high (TMB-high) and microsatellite instability (MSI) confirms their utility as immunotherapy biomarkers. ESMO Open, 7(1), 100336.
Pai, S. G., Carneiro, B. A., Chae, Y. K., et al. (2017). Correlation of tumor mutational burden and treatment outcomes in patients with colorectal cancer. Journal of Gastrointestinal Oncology, 8(5), 858–866.
Luo, J. H., Zhang, X. X., & Sun, W. H. (2022). F12 as a reliable diagnostic and prognostic biomarker associated with immune infiltration in papillary thyroid cancer. Aging, 14(8), 3687–3704.
Gazinska, P., Milton, C., Iacovacci, J., et al. (2022). Dynamic changes in the NK-, neutrophil-, and B-cell immunophenotypes relevant in high metastatic risk post neoadjuvant chemotherapy-resistant early breast cancers. Clinical Cancer Research : An Official Journal of the American Association for Cancer Research, 28(20), 4494–4508.
Li, H., Luo, F., Jiang, X., et al. (2022). CircITGB6 promotes ovarian cancer cisplatin resistance by resetting tumor-associated macrophage polarization toward the M2 phenotype. Journal for Immunotherapy of Cancer, 10(3), e004029.
Shi, Q., Shen, Q., Liu, Y., et al. (2022). Increased glucose metabolism in TAMs fuels O-GlcNAcylation of lysosomal Cathepsin B to promote cancer metastasis and chemoresistance. Cancer Cell, 40(10), 1207-1222.e10.
Zhang, J., Song, J., Tang, S., et al. (2023). Multi-omics analysis reveals the chemoresistance mechanism of proliferating tissue-resident macrophages in PDAC via metabolic adaptation. Cell Reports, 42(6), 112620.
Acknowledgements
We express our gratitude to all the databases mentioned in the article for granting us permission to utilize their data.
Funding
This study was funded by the Shandong Provincial Medical and Health Science and Technology Development Plan Project (No.202203100054).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Ethics Approval and Consent to Participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xing, L., Wu, S., Xue, S. et al. A Novel Neutrophil Extracellular Trap Signature Predicts Patient Chemotherapy Resistance and Prognosis in Lung Adenocarcinoma. Mol Biotechnol (2024). https://doi.org/10.1007/s12033-024-01170-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-024-01170-1