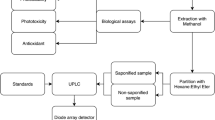
Abstract
Lysobacter is known as a bacterial genus with biotechnological potential, producing an array of enzymes, antimicrobial metabolites, and bioactive antioxidant compounds, including aryl polyene (APE) pigments that have been described as protecting substances against photooxidative damage and lipid peroxidation. In this study, the pigment extracted from keratinolytic Lysobacter sp. A03 isolated from Antarctic environment was characterized. The results of KOH test, UV–vis spectroscopy, CIELAB color system, 1H-NMR, and FTIR-ATR spectroscopy suggest the pigment is a yellow xanthomonadin-like pigment. The in vitro antioxidant activity of the pigment was confirmed by the scavenging of ABTS and DPPH radicals. In silico analysis of the genome through antiSMASH software was also performed and the secondary metabolite gene clusters for APE and resorcinol synthesis were identified, suggesting that proteins responsible for the pigment biosynthesis are encoded in Lysobacter A03 genome.





Similar content being viewed by others
Data Availabitily
The genome data from Lysobacter sp. strain A03 is publicy available at GenBank (https://www.ncbi.nlm.nih.gov/genbank/), accession number JXSS00000000.
References
Aruldass, C. A., Dufossé, L., & Ahmad, W. A. (2018). Current perspective of yellowish-orange pigments from microorganisms: A review. Journal of Cleaner Production, 180, 168–182. https://doi.org/10.1016/j.clepro.2018.01.093
Liu, J., Luo, Y., Guo, T., Tang, C., Chai, X., Zhao, W., Bai, J., & Lin, Q. (2020). Cost-effective pigment production by Monascus purpureus using rice straw hydrolysate as substrate in submerged fermentation. Journal of Bioscience and Bioengineering, 129, 229–236. https://doi.org/10.1016/j.jbiosc.2019.08.007
Venil, C. K., Dufossé, L., & Devi, P. R. (2020). Bacterial pigments: Sustainable compounds with market potential for pharma and food industry. Frontiers in Sustainable Food Systems, 4, 100. https://doi.org/10.3389/fsufs.2020.00100
Orlandi, V. T., Martegani, E., Giaroni, C., Baj, A., & Bolognese, F. (2022). Bacterial pigments: A colorful palette reservoir for biotechnological applications. Biotechnology and Applied Biochemistry, 69, 981–1001. https://doi.org/10.1002/bab.2170
Agarwal, H., Bajpai, S., Mishra, A., Kohli, I., Varma, A., Fouillaud, M., Dufossé, L., & Joshi, N. C. (2023). Bacterial pigments and their multifaceted roles in contemporary biotechnology and pharmacological applications. Microorganisms, 11, 614. https://doi.org/10.3390/microorganisms11030614
Rana, B., Bhattacharyya, M., Patni, B., Arya, M., & Joshi, G. K. (2021). The realm of microbial pigments in the food color market. Frontiers in Sustainable Food Systems, 5, 603892. https://doi.org/10.1189/fsufs.2021.603892
Pailliè-Jiménez, M. E., Stincone, P., & Brandelli, A. (2020). Natural pigments of microbial origin. Frontiers in Sustainable Food Systems, 4, 590439. https://doi.org/10.3389/fsufs.2020.590439
Schöner, T. A., Gassel, S., Osawa, A., Tobias, N. J., Okuno, Y., Sakakibara, Y., Shindo, K., Sandmann, G., & Bode, H. B. (2016). Aryl polyenes, a highly abundant class of bacterial natural products, are functionally related to antioxidative carotenoids. ChemBioChem, 17, 247–253. https://doi.org/10.1002/cbic.201500474
Wang, Y., Qian, G., Li, Y., Wang, Y., Wang, Y., Wright, S., Li, Y., Shen, Y., Liu, F., & Du, L. (2013). Biosynthetic mechanism for sunscreens of the biocontrol agent Lysobacter enzymogenes. PLoS ONE, 8, e66633. https://doi.org/10.1371/journal.pone.0066633
He, Y. W., Cao, X. Q., & Poplawsky, A. R. (2020). Chemical structure, biological roles, biosynthesis and regulation of the yellow xanthomonadin pigments in the phytopathogenic genus Xanthomonas. Molecular Plant-Microbe Interactions, 33, 705–714. https://doi.org/10.1094/MPMI-11-19-0326-CR
Madden, K. S., Laroche, B., David, S., Batsanov, A. S., Thompson, D., Knowles, J. P., & Whiting, A. (2018). Approaches to styrenyl building blocks for the synthesis of polyene xanthomonadin and its analogues. European Journal of Organic Chemistry, 2018, 5312–5322. https://doi.org/10.1002/ejoc.201800540
Kumar, S., Bansal, K., Patil, P. P., & Patil, P. B. (2019). Phylogenomics insights into order and families of Lysobacterales. Access Microbiology, 1, e000015. https://doi.org/10.1099/acmi.0.000015
Lee, S. Y., Kim, P. S., Sung, H., Hyun, D. W., & Bae, J. W. (2022). Lysobacter ciconiae sp. nov., and Lysobacter avium sp. nov., isolated from the faeces of an Oriental stork. Journal of Microbiology, 60, 469–477. https://doi.org/10.1007/s12275-022-1647-5
Xu, S., Zhang, Z., Xie, X., Shi, Y., Chai, A., Fan, T., Li, B., & Li, L. (2022). Comparative genomics provides insights into the potential biocontrol mechanism of two Lysobacter enzymogenes strains with distinct antagonistic activities. Frontiers in Microbiology, 13, 966986. https://doi.org/10.3389/fmicb.2022.966986
Drenker, C., El Mazouar, D., Bücker, G., Weißhaupt, S., Wienke, E., Koch, E., Kunz, S., Reineke, A., Rondot, Y., & Linkies, A. (2023). Characterization of a disease-suppressive isolate of Lysobacter enzymogenes with broad antagonistic activity against bacterial, oomycetal and fungal pathogens in different crops. Plants, 12, 682. https://doi.org/10.3390/plants12030682
Xie, Y., Wright, S., Shen, Y., & Du, L. (2012). Bioactive natural products from Lysobacter. Natural Product Reports, 29, 1277–1287. https://doi.org/10.1039/c2np20064c
Saini, D. K., Chakdar, H., Pabbi, S., & Shukla, P. (2020). Enhancing production of microalgal biopigments through metabolic and genetic engineering. Critical Reviews in Food Science and Nutrition, 60, 391–405. https://doi.org/10.1080/10408398.2018.1533518
Sen, T., Barrow, C. J., & Deshmukh, S. K. (2019). Microbial pigments in the food industry -Challenges and the way forward. Frontiers in Nutrition, 6, 7. https://doi.org/10.3389/fnut.2019.00007
Pereira, J. Q., Lopes, F. C., Petry, M. V., Medina, L. F. C., & Brandelli, A. (2014). Isolation of three novel Antarctic psychrotolerant feather-degrading bacteria and partial purification of keratinolytic enzyme from Lysobacter sp. A03. International Biodeterioration and Biodegradation, 88, 1–7. https://doi.org/10.1016/j.ibiod.2013.11.012
Pereira, J. Q., Ambosini, A., Passaglia, L. M., & Brandelli, A. (2017). A new cold-adapted serine peptidase from Antarctic Lysobacter sp. A03: Insights about enzyme activity at low temperatures. International Journal of Biological Macromolecules, 103, 854–862. https://doi.org/10.1016/j.ijbiomac.2017.05.142
Fautz, E., & Reichenbach, H. (1980). A simple test for flexirubin-type pigments. FEMS Microbiology Letters, 8, 87–91. https://doi.org/10.1111/j.1574-6968.1980.tb05056.x
Nenadis, N., Wang, L. F., Tsimidou, M., & Zhang, H. Y. (2004). Estimation of scavenging activity of phenolic compounds using the ABTS(*+) assay. Journal of Agricultural and Food Chemistry, 52, 4669–4674. https://doi.org/10.1021/jf0400056
Best, I., Casimiro-Gonzales, S., Portugal, A., Olivera-Montenegro, L., Aguilar, L., Muñoz, A. M., & Ramos-Escudero, F. (2020). Phytochemical screening and DPPH radical scavenging activity of three morphotypes of Mauritia flexuosa L.f. from Peru, and thermal stability of a milk-based beverage enriched with carotenoids from these fruits. Heliyon, 6, e05209. https://doi.org/10.1016/j.heliyon.2020.e05209
Pereira, J. Q., Ambrosini, A., Sant’Anna, F. H., Tadra-Sfeir, M., Faoro, H., Pedrosa, F. O., Souza, E. M., Brandelli, A., & Passaglia, L. M. (2015). Whole-genome shotgun sequence of the keratinolytic bacterium Lysobacter sp. A03, isolated from the Antarctic environment. Genome Announcements, 3, e00246-15. https://doi.org/10.1128/genomeA.00246-15
Blin, K., Shaw, S., Augustijn, H. E., Reitz, Z. L., Biermann, F., Alanjary, M., Fetter, A., Terlouw, B. R., Metcalf, W. W., Helfrich, E. J. N., van Wezel, G. P., Medema, M. H., & Weber, T. (2023). antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures, and visualization. Nucleic Acids Research, 51, W46–W50. https://doi.org/10.1093/nar/gkad344
Auch, A. F., Klenk, H. P., & Göker, M. (2012). Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Standards in Genomic Sciences, 2, 142–148. https://doi.org/10.4056/sigs.541628
Asnicar, F., Thomas, A. M., Beghini, F., Mengoni, C., Manara, S., Manghi, P., Zhu, Q., Bolzan, M., Cumbo, F., May, U., Sanders, J. G., Zolfo, M., Kopylova, E., Pasolli, E., Knight, R., Mirarab, S., Huttenhower, C., & Segata, N. (2020). Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nature Communications, 11, 2500. https://doi.org/10.1038/s41467-020-16366-7
Kimura, T., Fukuda, W., Sanada, T., & Imanaka, T. (2015). Characterization of water-soluble dark-brown pigment from Antarctic bacterium, Lysobacter oligotrophicus. Journal of Bioscience and Bioengineering, 120, 58–61. https://doi.org/10.1016/j.jbiosc.2014.11.020
Schöner, T. A., Fuchs, S. W., Reinhold-Hurek, B., & Bode, H. B. (2014). Identification and biosynthesis of a novel xanthomonadin-dialkylresorcinol-hybrid from Azoarcus sp. BH72. PLoS One, 9, e90922. https://doi.org/10.1371/journal.pone.0090922
Rohman, A., Windarsih, A., Lukitaningsih, E., Rafi, M., Betania, K., & Fadzillah, N. A. (2019). The use of FTIR and Raman spectroscopy in combination with chemometrics for analysis of biomolecules in biomedical fluids: A review. Biomedical Spectroscopy and Imaging, 8, 55–71. https://doi.org/10.3233/BSI-200189
Andrewes, A. G., Jenkins, C. L., Starr, M. P., Shepherd, J., & Hope, H. (1976). Structure of xanthomonadin I, a novel dibrominated aryl-polyene pigment produced by the bacterium Xanthomonas juglandis. Tetrahedron Letters, 45, 4023–4024.
Deng, K., Yang, Z., Luo, J., & Wu, K. (2018). Synthesis of aryl and heterocyclic polyenes and their activity in free radical scavenging. Journal of Chemical Research, 42, 129–132. https://doi.org/10.3184/174751918X15199929401287
Jiménez, M. E. P., Pinilla, C. M. B., Rodrigues, E., & Brandelli, A. (2019). Extraction and partial characterisation of antioxidant pigment produced by Chryseobacterium sp. kr6. Natural Products Research, 33, 1541–1549. https://doi.org/10.1080/14786419.2017.1423304
Li, F., Xue, F., & Yu, X. (2017). GC-MS, FTIR and Raman analysis of antioxidant components of red pigments from Stemphylium lycopersici. Current Microbiology, 74, 532–539. https://doi.org/10.1007/s00284-017-1220-3
Ahmad, W. A., Ahmad, W. Y. W., Zakaria, Z. A., & Yusof, N. Z. (2012). Isolation of pigment-producing bacteria and characterization of the extracted pigments. In W. A. Ahmad, W. Y. W. Ahmad, Z. A. Zakaria, & N. Z. Yusof (Eds.), Application of Bacterial Pigments as Colorant Springer Briefs in Molecular Science (pp. 22–44). Berlin: Springer. https://doi.org/10.1007/978-3-642-24520-6_2
Grammbitter, G. L. C., Shi, Y. M., Shi, Y. N., Vemulapalli, S. P. B., Richter, C., Schwalbe, H., Alanjary, M., Schüffler, A., Witt, M., Griesinger, C., & Bode, H. B. (2020). The chemical structures of widespread microbial aryl polyene lipids. bioRxiv. https://doi.org/10.1101/2020.12.19.423268
Aririatu, L. E., & Kester, A. S. (1985). Isolation and characterization of the pigment esters of Xanthomonas juglandis (campestris). Microbiology, 131, 2947–2052. https://doi.org/10.1099/00221289-131-9-2047
Campos, M., Gómez, K., Ordoñez, Y., & Ancona, D. (2013). Polyphenols, ascorbic acid and carotenoids contents and antioxidant properties of habanero pepper (Capsicum chinense) fruit. Food and Nutrition Sciences, 4, 47–54. https://doi.org/10.4236/fns.2013.48A006
Línzembold, I., Czett, D., Böddi, K., Kurtán, T., Király, S. B., Gulyás-Fekete, G., Takátsy, A., Lóránd, T., Deli, J., Agócs, A., & Nagy, V. (2020). Study on the synthesis, antioxidant properties, and self-assembly of carotenoid-flavonoid conjugates. Molecules, 25, 636. https://doi.org/10.3390/molecules25030636
Miller, N. J., Sampson, J., Candeias, L. P., Bramley, P. M., & Rice-Evans, C. A. (1996). Antioxidant activities of carotenes and xanthophylls. FEBS Letters, 384, 240–242. https://doi.org/10.1016/0014-5793(96)00323-7
Cimermancic, P., Medema, M. H., Claesen, J., Kurita, K., Wieland Brown, L. C., Mavrommatis, K., Pati, A., Godfrey, P. A., Koehrsen, M., Clardy, J., Birren, B. W., Takano, E., Sali, A., Linington, R. G., & Fischbach, M. A. (2014). Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell, 158, 412–421. https://doi.org/10.1016/j.cell.2014.06.034
Órdenes-Aenishanslins, N., Anziani-Ostuni, G., Vargas-Reyes, M., Alarcón, J., Tello, A., & Pérez-Donoso, J. M. (2016). Pigments from UV-resistant Antarctic bacteria as photosensitizers in dye sensitized solar cells. Journal of Photochemistry and Photobiology B, 162, 707–714. https://doi.org/10.1016/j.jphotobiol.2016.08.004
Goel, A. K., Rajagopal, L., Nagesh, N., & Sonti, R. V. (2002). Genetic locus encoding functions involved in biosynthesis and outer membrane localization of xanthomonadin in Xanthomonas oryzae pv. oryzae. Journal of Bacteriology, 184, 3539–3548. https://doi.org/10.1128/JB.184.13.3539-3548.2002
Mindrebo, J. T., Patel, A., Kim, W. E., Davis, T. D., Chen, A., Bartholow, T. G., La Clair, J. J., McCammon, J. A., Noel, J. P., & Burkart, M. D. (2020). Gating mechanism of elongating β-ketoacyl-ACP synthases. Nature Communications, 11, 1727. https://doi.org/10.1038/s41467-020-15455-x
de Bruijn, I., Cheng, X., de Jager, V., Expósito, R. G., Watrous, J., Patel, N., Postma, J., Dorrestein, P. C., Kobayashi, D., & Raaijmakers, J. M. (2015). Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genomics, 16, 991. https://doi.org/10.1186/s12864-015-2191-z
Acknowledgements
This work was supported by CNPq (grant 308880/2021-8) and CAPES (Brasilia, Brazil). MEPJ was a former recipient of a PhD fellowship from COLFUTURO (Bogotá, Colombia).
Funding
Funding was provided by Conselho Nacional de Desenvolvimento Científico e Tecnológico (Grant No: 308880/2021-8) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pailliè-Jiménez, M.E., Stincone, P., Pereira, J.Q. et al. Isolation and Characterization of an Antioxidant Aryl Polyene Pigment from Antarctic Bacterium Lysobacter sp. A03. Mol Biotechnol (2024). https://doi.org/10.1007/s12033-024-01132-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-024-01132-7