Abstract
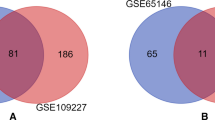
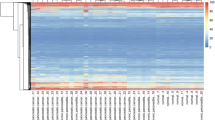
Acute pancreatitis (AP) and chronic pancreatitis (CP) are considered to be two separate pancreatic diseases in most studies, but some clinical retrospective analyses in recent years have found some degree of correlation between the two in actual treatment, however, the exact association is not clear. In this study, bioinformatics analysis was utilized to examine microarray sequencing data in mice, with the aim of elucidating the critical signaling pathways and genes involved in the progression from AP to CP. Differential gene expression analyses on murine transcriptomes were conducted using the R programming language and the R/Bioconductor package. Additionally, gene network analysis was performed using the STRING database to predict correlations among genes in the context of pancreatic diseases. Functional enrichment and gene ontology pathways common to both diseases were identified using Metascape. The hub genes were screened in the cytoscape algorithm, and the mRNA levels of the hub genes were verified in mice pancreatic tissues of AP and CP. Then the drugs corresponding to the hub genes were obtained in the drug-gene relationship. A set of hub genes, including Jun, Cd44, Epcam, Spp1, Anxa2, Hsp90aa1, and Cd9, were identified through analysis, demonstrating their pivotal roles in the progression from AP to CP. Notably, these genes were found to be enriched in the Helper T-cell factor (Th17) signaling pathway. Up-regulation of these genes in both AP and CP mouse models was validated through quantitative real-time polymerase chain reaction (qRT-PCR) results. The significance of the Th17 signaling pathway in the transition from AP to CP was underscored by our findings. Specifically, the essential genes driving this progression were identified as Jun, Cd44, Epcam, Spp1, Anxa2, Hsp90aa1, and Cd9. Crucial insights into the molecular mechanisms underlying pancreatitis progression were provided by this research, offering promising avenues for the development of targeted therapeutic interventions.





Similar content being viewed by others
Data availability
The data in this article was obtained online and can be obtained from publicly available databases. This manuscript is not applicable.
References
Beyer, G., Habtezion, A., Werner, J., et al. (2020). Chronic pancreatitis. Lancet, 396, 499–512.
Singh, V. K., Yadav, D., & Garg, P. K. (2019). Diagnosis and management of chronic pancreatitis: A review. JAMA, 322, 2422–2434.
Balázs, A., Balla, Z., Kui, B., et al. (2018). Ductal mucus obstruction and reduced fluid secretion are early defects in chronic pancreatitis. Frontiers in Physiology, 9, 632.
Zhang, Y., Zhang, W. Q., Liu, X. Y., et al. (2023). Immune cells and immune cell-targeted therapy in chronic pancreatitis. Frontiers in Oncology, 13, 1151103.
Glaubitz, J., Wilden, A., Golchert, J., et al. (2022). In mouse chronic pancreatitis CD25(+)FOXP3(+) regulatory T cells control pancreatic fibrosis by suppression of the type 2 immune response. Nature Communications, 13, 4502.
Mayerle, J., Sendler, M., Hegyi, E., et al. (2019). Genetics, cell biology, and pathophysiology of pancreatitis. Gastroenterology, 156, 1951-1968.e1951.
Sidhu, S., Pandhi, P., Malhotra, S., et al. (2011). Rosiglitazone promotes pancreatic regeneration in experimental model of acute pancreatitis. Fundamental & Clinical Pharmacology, 25, 237–247.
Paragomi, P., Spagnolo, D. M., Breze, C. R., et al. (2020). Introduction and validation of a novel acute pancreatitis digital tool: Interrogating large pooled data from 2 prospectively ascertained cohorts. Pancreas, 49, 1276–1282.
Yadav, D., O’Connell, M., & Papachristou, G. I. (2012). Natural history following the first attack of acute pancreatitis. American Journal of Gastroenterology, 107, 1096–1103.
Liu, J., Gao, M., Nipper, M., et al. (2019). Activation of the intrinsic fibroinflammatory program in adult pancreatic acinar cells triggered by Hippo signaling disruption. PLoS Biology, 17, e3000418.
Han, S. Y., Conwell, D. L., Diaz, P. T., et al. (2022). The deleterious effects of smoking on the development and progression of chronic pancreatitis. Pancreatology, 22, 683–687.
Norberg, K. J., Nania, S., Li, X., et al. (2018). RCAN1 is a marker of oxidative stress, induced in acute pancreatitis. Pancreatology, 18, 734–741.
Kowalik, A. S., Johnson, C. L., Chadi, S. A., et al. (2007). Mice lacking the transcription factor Mist1 exhibit an altered stress response and increased sensitivity to caerulein-induced pancreatitis. American Journal of Physiology Gastrointestinal and Liver Physiology, 292, G1123-1132.
Ulmasov, B., Oshima, K., Rodriguez, M. G., et al. (2013). Differences in the degree of cerulein-induced chronic pancreatitis in C57BL/6 mouse substrains lead to new insights in identification of potential risk factors in the development of chronic pancreatitis. American Journal of Pathology, 183, 692–708.
Yuan, Q., Ren, J., Chen, X., et al. (2022). Contributions and prognostic performances of m7G RNA regulators in pancreatic adenocarcinoma. Chinese Medical Journal (Engl), 135, 2101–2103.
Jin, S., & Hyun, T. K. (2020). Ectopic expression of production of anthocyanin pigment 1 (PAP1) improves the antioxidant and anti-melanogenic properties of ginseng (Panax ginseng C.A. Meyer) Hairy Roots. Antioxidants (Basel), 9, 922.
Zhou, Y., Zhou, B., Pache, L., et al. (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications, 10, 1523.
Alur, V. C., Raju, V., Vastrad, B., et al. (2019). Mining featured biomarkers linked with epithelial ovarian cancerbased on bioinformatics. Diagnostics (Basel), 9, 39.
Yang, X., Chen, J., Wang, J., et al. (2022). Very-low-density lipoprotein receptor-enhanced lipid metabolism in pancreatic stellate cells promotes pancreatic fibrosis. Immunity, 55, 1185-1199.e1188.
Shu, J., Ren, Y., Tan, W., et al. (2022). Identification of potential drug targets for vascular dementia and carotid plaques by analyzing underlying molecular signatures shared by them. Frontiers in Aging Neuroscience, 14, 967146.
Freshour, S. L., Kiwala, S., Cotto, K. C., et al. (2021). Integration of the drug-gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Research, 49, D1144-d1151.
Wen, L., Voronina, S., Javed, M. A., et al. (2015). Inhibitors of ORAI1 prevent cytosolic calcium-associated injury of human pancreatic acinar cells and acute pancreatitis in 3 mouse models. Gastroenterology, 149, 481-492.e487.
Bansod, S., Aslam Saifi, M., Khurana, A., et al. (2020). Nimbolide abrogates cerulein-induced chronic pancreatitis by modulating β-catenin/Smad in a sirtuin-dependent way. Pharmacological Research, 156, 104756.
Huang, Q. Y., Zhang, R., Zhang, Q. Y., et al. (2023). Disulfiram reduces the severity of mouse acute pancreatitis by inhibiting RIPK1-dependent acinar cell necrosis. Bioorganic Chemistry, 133, 106382.
Yasuda, K., Takeuchi, Y., & Hirota, K. (2019). The pathogenicity of Th17 cells in autoimmune diseases. Seminars in Immunopathology, 41, 283–297.
Littman, D. R., & Rudensky, A. Y. (2010). Th17 and regulatory T cells in mediating and restraining inflammation. Cell, 140, 845–858.
Griffith, M., Griffith, O. L., Coffman, A. C., et al. (2013). DGIdb—Mining the druggable genome. Nature Methods, 10, 1209–1210.
Freshour, S. L., Kiwala, S., Cotto, K. C., et al. (2020). Integration of the drug-gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Research, 49, D1144–D1151.
Ali, U. A., Issa, Y., Hagenaars, J. C., et al. (2016). Risk of recurrent pancreatitis and progression to chronic pancreatitis after a first episode of acute pancreatitis—ScienceDirect. Clinical Gastroenterology and Hepatology, 14, 738–746.
Alexandre, M., Pandol, S. J., Gorelick, F. S., et al. (2011). The emerging role of smoking in the development of pancreatitis. Pancreatology, 11, 469–474.
Yadav, D., Hawes, R. H., Brand, R. E., et al. (2009). Alcohol consumption, cigarette smoking, and the risk of recurrent acute and chronic pancreatitis. Archives of Internal Medicine, 169, 1035–1045.
Li, C., Heidt, D. G., Dalerba, P., et al. (2007). Identification of pancreatic cancer stem cells. Cancer Research, 67, 1030–1037.
Wang, V. M., Ferreira, R. M. M., Almagro, J., et al. (2019). CD9 identifies pancreatic cancer stem cells and modulates glutamine metabolism to fuel tumour growth. Nature Cell Biology, 21, 1425–1435.
Chen, K., Wang, Q., Li, M., et al. (2021). Single-cell RNA-seq reveals dynamic change in tumor microenvironment during pancreatic ductal adenocarcinoma malignant progression. eBioMedicine, 66, 103315.
Singhi, A. D., Pai, R. K., Kant, J. A., et al. (2014). The histopathology of PRSS1 hereditary pancreatitis. American Journal of Surgical Pathology, 38, 346–353.
Singhi, A. D., Koay, E. J., Chari, S. T., et al. (2019). Early detection of pancreatic cancer: Opportunities and challenges. Gastroenterology, 156, 2024–2040.
Feng, Y., Li, W., Wang, Z., et al. (2022). The p-STAT3/ANXA2 axis promotes caspase-1-mediated hepatocyte pyroptosis in non-alcoholic steatohepatitis. Journal of Translational Medicine, 20, 497.
Wang, K., Zhang, T., Lei, Y., et al. (2018). Identification of ANXA2 (annexin A2) as a specific bleomycin target to induce pulmonary fibrosis by impeding TFEB-mediated autophagic flux. Autophagy, 14, 269–282.
Hendley, A. M., Rao, A. A., Leonhardt, L., et al. (2021). Single-cell transcriptome analysis defines heterogeneity of the murine pancreatic ductal tree. eLife, 10, e677776.
Adams, C. R., Htwe, H. H., Marsh, T., et al. (2019). Transcriptional control of subtype switching ensures adaptation and growth of pancreatic cancer. eLife, 8, e45313.
Park, H., Li, Z., Yang, X. O., et al. (2005). A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nature Immunology, 6, 1133–1141.
Cen, M. E., Wang, F., Su, Y., et al. (2018). Gastrointestinal microecology: A crucial and potential target in acute pancreatitis. Apoptosis, 23, 377–387.
Cosmi, L., Santarlasci, V., Maggi, L., et al. (2014). Th17 plasticity: Pathophysiology and treatment of chronic inflammatory disorders. Current Opinion in Pharmacology, 17, 12–16.
Guo, J., Li, Z., Tang, D., et al. (2020). Th17/Treg imbalance in patients with severe acute pancreatitis: Attenuated by high-volume hemofiltration treatment. Medicine (Baltimore), 99, e21491.
Campbell, C., Dikiy, S., Bhattarai, S. K., et al. (2018). Extrathymically generated regulatory T cells establish a niche for intestinal border-dwelling bacteria and affect physiologic metabolite balance. Immunity, 48, 1245-1257.e1249.
Lee, B., Jones, E. K., Manohar, M., et al. (2023). Distinct serum immune profiles define the spectrum of acute and chronic pancreatitis from the multicenter prospective evaluation of chronic pancreatitis for epidemiologic and translational studies (PROCEED) study. Gastroenterology, 165, 173–186.
Zhao, Q., Manohar, M., Wei, Y., et al. (2019). STING signalling protects against chronic pancreatitis by modulating Th17 response. Gut, 68, 1827–1837.
Funding
This work was supported by National Natural Science Foundation of China (No. 82100684), Natural Science Foundation of Chongqing China (CSTB2022NSCQ-MSX1493), Scientific Research Foundation of Chongqing University of Technology, National Natural Science Foundation Incubation project of Chongqing University of Technology (2022PYZ037), and the Funding Achievements of the Action Plan for High Quality Development of Graduate Education at Chongqing University of Technology (gzlcx20233384).
Author information
Authors and Affiliations
Contributions
Yan Shen designed this study and critically revised the manuscript. Lu Yuan and Yiyuan Liu drafted the manuscript. Lingyan Fan acquired and analyzed the data. Yiyuan Liu, Lu Yuan, Cai Sun and Sha Ran did literature search and performed the experiments. Kuilong Huang directed the qRT-PCR experiments and analyzed the data. All authors have reviewed and confirmed the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors confirm that they do not have any acknowledged competing financial interests or personal affiliations that might have appeared to influence the research documented in this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yuan, L., Liu, Y., Fan, L. et al. Identification of Potential Hub Genes Related to Acute Pancreatitis and Chronic Pancreatitis via Integrated Bioinformatics Analysis and In Vitro Analysis. Mol Biotechnol (2024). https://doi.org/10.1007/s12033-024-01118-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-024-01118-5