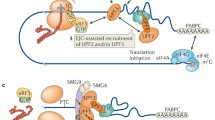
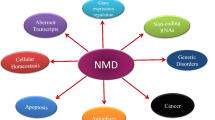
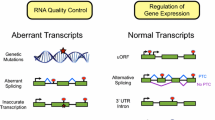
Abstract
Nonsense-mediated mRNA decay (NMD) is an evolutionarily conserved surveillance mechanism in eukaryotes primarily deployed to ensure RNA quality control by eliminating aberrant transcripts and also involved in modulating the expression of several physiological transcripts. NMD, the mRNA surveillance pathway, is a major form of gene regulation in eukaryotes. NMD serves as one of the most significant quality control mechanisms as it primarily scans the newly synthesized transcripts and differentiates the aberrant and non-aberrant transcripts. The synthesis of truncated proteins is restricted, which would otherwise lead to cellular dysfunctions. The up-frameshift factors (UPFs) play a central role in executing the NMD event, largely by recognizing and recruiting multiple protein factors that result in the decay of non-physiological mRNAs. NMD exhibits astounding variability in its ability across eukaryotes in an array of pathological and physiological contexts. The detailed understanding of NMD and the underlying molecular mechanisms remains blurred. This review outlines our current understanding of NMD, in regulating multifaceted cellular events during development and disease. It also attempts to identify unanswered questions that deserve further investigation.
Graphical Abstract





Similar content being viewed by others
Data Availability
Not applicable.
Abbreviations
- NMD:
-
Nonsense-mediated mRNA decay
- PTC:
-
Premature termination codons
- PABPC1:
-
PolyA-binding protein C1
- UPF:
-
Up-frameshift protein
- EJC:
-
Exon junction complexes
- 3′UTR:
-
3′ Untranslated regions
- SMG:
-
Suppressor of Morphogenesis in Genitalia
- ORF:
-
Open reading frame
- UPF1LL:
-
UPF1’s long loop
- MIF4G-3:
-
Middle domain of eukaryotic initiation factor 4G (eIF4G)
- RRM:
-
RNA Recognition Motif
- EBM:
-
EJC-binding motif
- RBM8A:
-
RNA-binding protein 8A
- CASC3:
-
Cancer susceptibility candidate gene 3
- RNPS1:
-
RNA-binding protein with serine-rich domain 1
- DECID:
-
Decay-inducing complex
- SR proteins:
-
Serine and arginine-rich proteins
- mRNPs:
-
MRNA molecules with ribonucleoproteins
References
Mendell, J. T., Sharifi, N. A., Meyers, J. L., Martinez-Murillo, F., & Dietz, H. C. (2004). Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nature genetics, 36(10), 1073–1078.
Wittmann, J., Hol, E. M., & Jäck, H. M. (2006). hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Molecular and cellular biology, 26(4), 1272–1287.
Hwang, H. J., Park, Y., & Kim, Y. K. (2021). UPF1: From mRNA surveillance to protein quality control. Biomedicines, 9(8), 995.
Karousis, E. D., Nasif, S., & Mühlemann, O. (2016). Nonsense-mediated mRNA decay: Novel mechanistic insights and biological impact. Wiley Interdisciplinary Reviews: RNA, 7(5), 661–682.
Nogueira, G., Fernandes, R., García-Moreno, J. F., & Romão, L. (2021). Nonsense-mediated RNA decay and its bipolar function in cancer. Molecular Cancer, 20(1), 1–19.
Popp, M. W., & Maquat, L. E. (2018). Nonsense-mediated mRNA decay and cancer. Current Opinion in Genetics & Development, 48, 44–50.
Ishigaki, Y., Li, X., Serin, G., & Maquat, L. E. (2001). Evidence for a pioneer round of mRNA translation: MRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell, 106(5), 607–617.
Eberle, A. B., Lykke-Andersen, S., Mühlemann, O., & Jensen, T. H. (2009). SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nature Structural & Molecular Biology, 16(1), 49–55.
Huntzinger, E., Kashima, I., Fauser, M., Saulière, J., & Izaurralde, E. (2008). SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA, 14(12), 2609–2617.
Patro, I., Sahoo, A., Nayak, B. R., Das, R., Majumder, S., & Panigrahi, G. K. (2023). Nonsense-mediated mRNA decay: Mechanistic insights and physiological significance. Molecular Biotechnology. https://doi.org/10.1007/s12033-023-00927-4
Buhler, M., Steiner, S., Mohn, F., Paillusson, A., & Muhlemann, O. (2006). EJC-independent degradation of nonsense immunoglobulin-mumRNA depends on 3′ UTR length. Nature Structural & Molecular Biology, 13, 462–464.
Singh, G., Rebbapragada, I., & Lykke-Andersen, J. (2008). A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biology, 6(4), e111.
Medghalchi, S. M., Frischmeyer, P. A., Mendell, J. T., Kelly, A. G., Lawler, A. M., & Dietz, H. C. (2001). Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Human Molecular Genetics, 10(2), 99–105.
Xie, N., Shen, G., Gao, W., Huang, Z., Huang, C., & Fu, L. (2023). Neoantigens: Promising targets for cancer therapy. Signal Transduction and Targeted Therapy, 8(1), 9.
Wittkopp, N., Huntzinger, E., Weiler, C., Saulière, J., Schmidt, S., Sonawane, M., & Izaurralde, E. (2009). Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Molecular and Cellular Biology, 29(13), 3517–3528.
Panigrahi, G. K., & Satapathy, K. B. (2020). Arabidopsis DCP5, a decapping complex protein interacts with Ubiquitin-5 in the processing bodies. Plant Archives, 20(1), 2243–2247.
Valcarce, D. G., Riesco, M. F., Cuesta-Martín, L., Esteve-Codina, A., Martínez-Vázquez, J. M., & Robles, V. (2023). Stress decreases spermatozoa quality and induces molecular alterations in zebrafish progeny. BMC Biology, 21(1), 1–20.
Conti, E., & Izaurralde, E. (2005). Nonsense-mediated mRNA decay: Molecular insights and mechanistic variations across species. Current Opinion in Cell Biology, 17, 316–325.
Mühlemann, O. (2008). Recognition of nonsense mRNA: Towards a unified model. Biochemical Society Transactions, 36, 497–501.
He, F., & Jacobson, A. (2015). Nonsense-mediated mRNA decay: Degradation of defective transcripts is only part of the story. Annual Review of Genetics, 49, 339–366.
Gehring, N. H., Kunz, J. B., Neu-Yilik, G., Breit, S., Viegas, M. H., Hentze, M. W., & Kulozik, A. E. (2005). Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Molecular Cell, 20(1), 65–75.
Tarpey, P., Raymond, F. L., Nguyen, L. S., Rodriguez, J., Hackett, A., Vandeleur, L., Smith, R., Shoubridge, C., Edkins, S., Stevens, C., O’Meara, S., Tofts, C., Barthorpe, S., Buck, G., Cole, J., Halliday, K., Hills, K., Jones, D. R., Mironenko, T., … Gécz, J. (2007). Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nature Genetics, 39(9), 1127–1133.
Bao, J., Vitting-Seerup, K., Waage, J., Tang, C., Ge, Y., Porse, B. T., & Yan, W. (2016). UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3′UTR transcripts. PLoS Genetics, 12(5), e1005863.
Ge, Z., Quek, B. L., Beemon, K. L., & Hogg, J. R. (2016). Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. eLife, 5, e11155.
Zhang, Z., & Krainer, A. R. (2004). Involvement of SR proteins in mRNA surveillance. Molecular Cell, 16(4), 597–607.
Kurihara, Y., Makita, Y., Kawauchi, M., Kageyama, A., Kuriyama, T., & Matsui, M. (2022). Intergenic splicing-stimulated transcriptional readthrough is suppressed by nonsense-mediated mRNA decay in Arabidopsis. Communications Biology, 5, 1390. https://doi.org/10.1038/s42003-022-04348-y
Zünd, D., Gruber, A. R., Zavolan, M., & Mühlemann, O. (2013). Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nature Structural & Molecular Biology, 20(8), 936–943.
Kurosaki, T., & Maquat, L. E. (2013). Rules that govern UPF1 binding to mRNA 3′ UTRs. Proceedings of the National Academy of Sciences, 110(9), 3357–3362.
Hurt, J. A., Robertson, A. D., & Burge, C. B. (2013). Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Research, 23(10), 1636–1650.
Chakrabarti, S., Jayachandran, U., Bonneau, F., Fiorini, F., Basquin, C., Domcke, S., Hir, H. L., & Conti, E. (2011). Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Molecular Cell, 41(6), 693–703.
Kadlec, J., Guilligay, D., Ravelli, R. B., & Cusack, S. (2006). Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA, 12, 1817–1824.
Cheng, Z., Muhlrad, D., Lim, M. K., Parker, R., & Song, H. (2007). Structural and functional insights into the human Upf1 helicase core. EMBO Journal, 26, 253–264.
Clerici, M., Mourão, A., Gutsche, I., Gehring, N. H., Hentze, M. W., & Kulozik, A. (2009). Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO Journal, 28, 2293–2306.
Gowravaram, M., Bonneau, F., Kanaan, J., Maciej, V. D., Fiorini, F., & Raj, S. (2018). A conserved structural element in the RNA helicase UPF1 regulates its catalytic activity in an isoform-specific manner. Nucleic Acids Research, 46, 2648–2659.
Durand, S., Franks, T. M., & Lykke-Andersen, J. (2016). Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nature Communications, 7, 12434.
Okada-Katsuhata, Y., Yamashita, A., Kutsuzawa, K., Izumi, N., Hirahara, F., & Ohno, S. (2012). N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Research, 40, 1251–1266.
Chakrabarti, S., Bonneau, F., Schüssler, S., Eppinger, E., & Conti, E. (2014). Phospho-dependent and phospho-independent interactions of the helicase UPF1 with the NMD factors SMG5-SMG7 and SMG6. Nucleic Acids Research, 42, 9447–9460.
Nicholson, P., Josi, C., Kurosawa, H., Yamashita, A., & Mühlemann, O. (2014). A novel phosphorylation-independent interaction between SMG6 and UPF1 is essential for human NMD. Nucleic Acids Research, 42, 9217–9235.
Feng, Q., Jagannathan, S., & Bradley, R. K. (2017). The RNA surveillance factor UPF1 represses myogenesis via its E3 ubiquitin ligase activity. Molecular Cell, 67, 239-251.e236.
Joazeiro, C. A. P. (2019). Mechanisms and functions of ribosome-associated protein quality control. Nature Reviews Molecular Cell Biology, 20, 368–383.
Powers, K. T., Szeto, J. A., & Schaffitzel, C. (2020). New insights into no-go, non-stop and nonsense-mediated mRNA decay complexes. Current Opinion in Structural Biology, 65, 110–118.
Inglis, A.J., Guna, A., Merchán, Á.G, Pal, A., Esantsi, T.K., Keys, H.R. (2022). Coupled protein quality control during nonsense mediated mRNA decay. bioRxiv
Chamieh, H., Ballut, L., Bonneau, F., & Le Hir, H. (2008). NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nature Structural & Molecular Biology, 15, 85–93.
Xin, M. A., Yan, L. I., Chengyan, C. H. E. N., Yanmin, S. H. E. N., Hua, W. A. N. G., & Tangliang, L. I. (2023). Spatial expression of the nonsense-mediated mRNA decay factors UPF3A and UPF3B among mouse tissues. Journal of Zhejiang University. Science. B, 24(11), 1062.
Kishor, A., Ge, Z., & Hogg, J. R. (2019). hnRNP L-dependent protection of normal mRNAs from NMD subverts quality control in B cell lymphoma. EMBO Journal, 38, e99128.
Kishor, A., Fritz, S. E., Haque, N., Ge, Z., Tunc, I., & Yang, W. (2020). Activation and inhibition of nonsense-mediated mRNA decay control the abundance of alternative polyadenylation products. Nucleic Acids Research, 48, 7468–7482.
Fritz, S. E., Ranganathan, S., Wang, C. D., & Hogg, J. R. (2022). An alternative UPF1 isoform drives conditional remodeling of nonsense-mediated mRNA decay. EMBO Journal, 41, e108898.
Mabin, J. W., Woodward, L. A., Patton, R. D., Yi, Z., Jia, M., Wysocki, V. H., Bundschuh, R., & Singh, G. (2018). The exon junction complex undergoes a compositional switch that alters mRNP structure and nonsense-mediated mRNA decay activity. Cell Reports, 25(9), 2431–2446.
Gerbracht, J. V., Boehm, V., Britto-Borges, T., Kallabis, S., Wiederstein, J. L., Ciriello, S., Aschemeier, D. U., Krüger, M., Frese, C. K., Altmüller, J., Dieterich, C., & Gehring, N. H. (2020). CASC3 promotes transcriptome-wide activation of nonsense-mediated decay by the exon junction complex. Nucleic Acids Research, 48(15), 8626–8644.
Kadlec, J., Izaurralde, E., & Cusack, S. (2004). The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nature Structural & Molecular Biology, 11, 330–337.
Panigrahi, G. K., Sahoo, A., & Satapathy, K. B. (2021). Insights to plant immunity: Defense signaling to epigenetics. Physiological and Molecular Plant Pathology, 113, 1–7.
Melero, R., Buchwald, G., Castaño, R., Raabe, M., Gil, D., Lázaro, M., Urlaub, H., Conti, E., & Llorca, O. (2012). The cryo-EM structure of the UPF–EJC complex shows UPF1 poised toward the RNA 3′ end. Nature Structural & Molecular Biology, 19(5), 498–505.
Sukarta, O. C. A., Slootweg, E. J., & Goverse, A. (2016). Structure informed insights for NLR functioning in plant immunity. Seminars in Cell & Developmental Biology, 56, 134–149.
Buchwald, G., Ebert, J., Basquin, C., Sauliere, J., Jayachandran, U., Bono, F., Hir, H. L., & Conti, E. (2010). Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proceedings of the National Academy of Sciences, 107(22), 10050–10055.
Kashima, I., Yamashita, A., Izumi, N., Kataoka, N., Morishita, R., Hoshino, S., Ohno, M., Dreyfuss, G., & Ohno, S. (2006). Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & Development, 20(3), 355–367.
Bono, F., Ebert, J., Lorentzen, E., & Conti, E. (2006). The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell, 126(4), 713–725.
Andersen, C. B. F., Ballut, L., Johansen, J. S., Chamieh, H., Nielsen, K. H., Oliveira, C. L. P., Pedersen, J. S., Séraphin, B., Hir, H. L., & Andersen, G. R. (2006). Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science, 313(5795), 1968–1972.
Panigrahi, G. K., Sahoo, A., & Satapathy, K. B. (2021). Differential expression of selected Arabidopsis resistant genes under abiotic stress conditions. Plant Science Today, 8(4), 859–864.
Shivaprasad, P. V., Chen, H. M., Patel, K., Bond, D. M., Santos, B. A., & Baulcombe, D. C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. The Plant Cell, 24, 859–874.
Singh, G., Kucukural, A., Cenik, C., Leszyk, J. D., Shaffer, S. A., Weng, Z., & Moore, M. J. (2012). The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell, 151(4), 750–764.
Nguyen, L. S., Jolly, L., Shoubridge, C., Chan, W. K., Huang, L., & Laumonnier, F. (2012). Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Molecular Psychiatry, 17, 1103–1115.
Sato, H., Hosoda, N., & Maquat, L. E. (2008). Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Molecular Cell, 29(2), 255–262.
Wallmeroth, D., Lackmann, J. W., Kueckelmann, S., Altmüller, J., Dieterich, C., Boehm, V., & Gehring, N. H. (2022). Human UPF3A and UPF3B enable fault-tolerant activation of nonsense-mediated mRNA decay. The EMBO Journal, 41(10), e109191.
Yi, Z., Arvola, R. M., Myers, S., Dilsavor, C. N., Abu Alhasan, R., & Carter, B. N. (2022). Mammalian UPF3A and UPF3B can activate nonsense mediated mRNA decay independently of their exon junction complex binding. EMBO Journal, 41, e109202.
Kurihara, Y., Matsui, A., Hanada, K., Kawashima, M., Ishida, J., Morosawa, T., Tanaka, M., Kaminuma, E., Mochizuki, Y., Matsushima, A., Toyoda, T., Shinozaki, K., & Seki, M. (2009). Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proceedings of the National Academy of Sciences, 106(7), 2453–2458.
Viphakone, N., Sudbery, I., Griffith, L., Heath, C. G., Sims, D., & Wilson, S. A. (2019). Co-transcriptional loading of RNA export factors shapes the human transcriptome. Molecular Cell, 75, 310-323.e318.
Linder, P., & Jankowsky, E. (2011). From unwinding to clamping—The DEAD box RNA helicase family. Nature Reviews Molecular Cell Biology, 12, 505–516.
Andersen, C. B., Ballut, L., Johansen, J. S., Chamieh, H., Nielsen, K. H., & Oliveira, C. L. (2006). Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science, 313, 1968–1972.
Karasov, T. L., Chae, E., Herman, J. J., & Bergelson, J. (2017). Mechanisms to mitigate the trade-off between growth and defense. The Plant Cell, 29, 666–680.
Nielsen, K. H., Chamieh, H., Andersen, C. B., Fredslund, F., Hamborg, K., & Le Hir, H. (2009). Mechanism of ATP turnover inhibition in the EJC. RNA, 15, 67–75.
Le Hir, H., Izaurralde, E., Maquat, L. E., & Moore, M. J. (2000). The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO Journal, 19, 6860–6869.
Fribourg, S., Gatfield, D., Izaurralde, E., & Conti, E. (2003). A novel mode of RBD-protein recognition in the Y14-Mago complex. Natural Structural Biology, 10, 433–439.
Ballut, L., Marchadier, B., Baguet, A., Tomasetto, C., Séraphin, B., & Le Hir, H. (2005). The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nature Structural & Molecular Biology, 12, 861–869.
Sahoo, A., & Satapathy, K. B. (2021). Differential expression of Arabidopsis EJC core proteins under short-day and long-day growth conditions. Plant Science Today, 8(4), 815–819.
Kashima, I., Jonas, S., Jayachandran, U., Buchwald, G., Conti, E., & Lupas, A. N. (2010). SMG6 interacts with the exon junction complex via two conserved EJC-binding motifs (EBMs) required for nonsense-mediated mRNA decay. Genes & Development, 24, 2440–2450.
Garcia, D., Garcia, S., & Voinnet, O. (2014). Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host & Microbe, 16, 391–402.
Lykke-Andersen, J., Shu, M. D., & Steitz, J. A. (2001). Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science, 293, 1836–1839.
Gehring, N. H., Lamprinaki, S., Hentze, M. W., & Kulozik, A. E. (2009). The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biology, 7, e1000120.
Sakashita, E., Tatsumi, S., Werner, D., Endo, H., & Mayeda, A. (2004). Human RNPS1 and its associated factors: A versatile alternative pre-mRNA splicing regulator in vivo. Molecular and Cellular Biology, 24, 1174–1187.
Imseng, S., Aylett, C. H., & Maier, T. (2018). Architecture and activation of phosphatidylinositol 3-kinase related kinases. Current Opinion in Structural Biology, 49, 177–189.
Yamashita, A., Ohnishi, T., Kashima, I., Taya, Y., & Ohno, S. (2001). Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes & Development, 15, 2215–2228.
Brumbaugh, K. M., Otterness, D. M., Geisen, C., Oliveira, V., Brognard, J., & Li, X. (2004). The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Molecular Cell, 14, 585–598.
Yamashita, A., Izumi, N., Kashima, I., Ohnishi, T., Saari, B., & Katsuhata, Y. (2009). SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes & Development, 23, 1091–1105.
Arias-Palomo, E., Yamashita, A., Fernández, I. S., Núñez-Ramírez, R., Bamba, Y., & Izumi, N. (2011). The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes & Development, 25, 153–164.
Deniaud, A., Karuppasamy, M., Bock, T., Masiulis, S., Huard, K., & Garzoni, F. (2015). A network of SMG-8, SMG-9 and SMG-1 C-terminal insertion domain regulates UPF1 substrate recruitment and phosphorylation. Nucleic Acids Research, 43, 7600–7611.
Zhu, L., Li, L., Qi, Y., Yu, Z., & Xu, Y. (2019). Cryo-EM structure of SMG1-SMG8-SMG9 complex. Cell Research, 29, 1027–1034.
Li, L., Lingaraju, M., Basquin, C., Basquin, J., & Conti, E. (2017). Structure of a SMG8-SMG9 complex identifies a G-domain heterodimer in the NMD effector proteins. RNA, 23, 1028–1034.
Langer, L. M., Bonneau, F., Gat, Y., & Conti, E. (2021). Cryo-EM reconstructions of inhibitor-bound SMG1 kinase reveal an autoinhibitory state dependent on SMG8. eLife, 10, e72353.
Ohnishi, T., Yamashita, A., Kashima, I., Schell, T., Anders, K. R., & Grimson, A. (2003). Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Molecular Cell, 12, 1187–1200.
Kurosaki, T., Li, W., Hoque, M., Popp, M. W., Ermolenko, D. N., & Tian, B. (2014). A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes & Development, 28, 1900–1916.
Jonas, S., Weichenrieder, O., & Izaurralde, E. (2013). An unusual arrangement of two 14-3-3-like domains in the SMG5-SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. Genes & Development, 27, 211–225.
Fukuhara, N., Ebert, J., Unterholzner, L., Lindner, D., Izaurralde, E., & Conti, E. (2005). SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Molecular Cell, 17, 537–547.
Huntzinger, E., Kashima, I., Fauser, M., Saulière, J., & Izaurralde, E. (2008). SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA, 14, 2609–2617.
Loh, B., Jonas, S., & Izaurralde, E. (2013). The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes & Development, 27, 2125–2138.
Glavan, F., Behm-Ansmant, I., Izaurralde, E., & Conti, E. (2006). Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO Journal, 25, 5117–5125.
Eberle, A. B., Lykke-Andersen, S., Mühlemann, O., & Jensen, T. H. (2009). SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nature Structural & Molecular Biology, 16, 49–55.
Boehm, V., Haberman, N., Ottens, F., Ule, J., & Gehring, N. H. (2014). 3′ UTR length and messenger ribonucleoprotein composition determine endocleavage efficiencies at termination codons. Cell Reports, 9(2), 555–568.
Lykke-Andersen, S., Chen, Y., Ardal, B. R., Lilje, B., Waage, J., & Sandelin, A. (2014). Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes & Development, 28, 2498–2517.
Gatfield, D., & Izaurralde, E. (2004). Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature, 429, 575–578.
Schmidt, S. A., Foley, P. L., Jeong, D. H., Rymarquis, L. A., Doyle, F., & Tenenbaum, S. A. (2015). Identification of SMG6 cleavage sites and a preferred RNA cleavage motif by global analysis of endogenous NMD targets in human cells. Nucleic Acids Research, 43, 309–323.
Boehm, V., Kueckelmann, S., Gerbracht, J. V., Kallabis, S., Britto-Borges, T., & Altmüller, J. (2021). SMG5-SMG7 authorize nonsense-mediated mRNA decay by enabling SMG6 endonucleolytic activity. Nature Communications, 12, 3965.
Metze, S., Herzog, V. A., Ruepp, M. D., & Mühlemann, O. (2013). Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA, 19, 1432–1448.
Colombo, M., Karousis, E. D., Bourquin, J., Bruggmann, R., & Mühlemann, O. (2017). Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA, 23, 189–201.
Serdar, L. D., Whiteside, D. L., Nock, S. L., McGrath, D., & Baker, K. E. (2020). Inhibition of post-termination ribosome recycling at premature termination codons in UPF1 ATPase mutants. eLife, 9, e57834.
Lejeune, F., Li, X., & Maquat, L. E. (2003). Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Molecular Cell, 12, 675–687.
Chen, C. Y., & Shyu, A. B. (2003). Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Molecular and Cellular Biology, 23, 4805–4813.
Monaghan, L., Longman, D., & Cáceres, J. F. (2023). Translation-coupled mRNA quality control mechanisms. The EMBO Journal, 42(19), e114378.
Hogg, J. R., & Goff, S. P. (2010). Upf1 senses 3′ UTR length to potentiate mRNA decay. Cell, 143(3), 379–389.
Hosoda, N., Kim, Y. K., Lejeune, F., & Maquat, L. E. (2005). CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nature Structural & Molecular Biology, 12, 893–901.
Neu-Yilik, G., Raimondeau, E., Eliseev, B., Yeramala, L., Amthor, B., Deniaud, A., Huard, K., Kerschgens, K., Hentze, M. W., Schaffitzel, C., & Kulozik, A. E. (2017). Dual function of UPF3B in early and late translation termination. The EMBO Journal, 36(20), 2968–2986.
Franks, T. M., Singh, G., & Lykke-Andersen, J. (2010). Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsensemediated mRNA decay. Cell, 143, 938–950.
Eberle, A. B., Stalder, L., Mathys, H., Orozco, R. Z., & Muhlemann, O. (2008). Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biology, 6, e92.
Panigrahi, G. K., & Satapathy, K. B. (2020). Formation of Arabidopsis Poly(A)-Specific Ribonuclease associated processing bodies in response to pathogenic infection. Plant Archives, 20(2), 4907–4912.
Shigeoka, T., Kato, S., Kawaichi, M., & Ishida, Y. (2012). Evidence that the Upf1-related molecular motor scans the 3′-UTR to ensure mRNA integrity. Nucleic Acids Research, 40, 6887–6897.
Hogg, J. R., & Goff, S. P. (2010). Upf1 senses 3′UTR length to potentiate mRNA decay. Cell, 143, 379–389.
Lykke-Andersen, S., & Jensen, T. H. (2015). Nonsensemediated mRNA decay: An intricate machinery that shapes transcriptomes. Nature Reviews Molecular Cell Biology, 16, 665–677.
Kurosaki, T., Popp, M. W., & Maquat, L. E. (2019). Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nature Reviews Molecular Cell Biology, 20(7), 406–420.
Huang, L., & Wilkinson, M. F. (2012). Regulation of nonsensemediated mRNA decay. Wiley Interdiscip. Rev. RNA, 3, 807–828.
Boehm, V., & Gehring, N. H. (2016). Exon junction complexes: Supervising the gene expression assembly line. Trends in Genetics, 32, 724–735.
Woodward, L. A., Mabin, J. W., Gangras, P., & Singh, G. (2017). The exon junction complex: A lifelong guardian of mRNA fate. Wiley Interdisciplinary Reviews: RNA, 8(3), e1411.
Hir, H. L., Saulière, J., & Wang, Z. (2016). The exon junction complex as a node of post-transcriptional networks. Nature Reviews Molecular Cell Biology, 17(1), 41–54.
Lindeboom, R. G., Supek, F., & Lehner, B. (2016). The rules and impact of nonsense-mediated mRNA decay in human cancers. Nature Genetics, 48(10), 1112–1118.
Gangras, P., Gallagher, T. L., Parthun, M. A., Yi, Z., Patton, R. D., Tietz, K. T., Deans, N. C., Bundschuh, R., Amacher, S. L., & Singh, G. (2020). Zebrafish rbm8a and magoh mutants reveal EJC developmental functions and new 3′UTR intron-containing NMD targets. PLOS Genetics, 16(6), e1008830.
Silver, D. L., Watkins-Chow, D. E., Schreck, K. C., Pierfelice, T. J., Larson, D. M., Burnetti, A. J., Liaw, H.-J., Myung, K., Walsh, C. A., Gaiano, N., & Pavan, W. J. (2010). The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nature Neuroscience, 13(5), 551–558.
McMahon, J. J., Miller, E. E., & Silver, D. L. (2016). The exon junction complex in neural development and neurodevelopmental disease. International Journal of Developmental Neuroscience, 55, 117–123.
Hoek, T. A., Khuperkar, D., Lindeboom, R. G. H., Stijn Sonneveld, B. M. P., Verhagen, S. B., Vermeulen, M., & Tanenbaum, M. E. (2019). Single-molecule imaging uncovers rules governing nonsense-mediated mRNA decay. Molecular Cell, 75(2), 324–339.
Nott, A., Le Hir, H., & Moore, M. J. (2004). Splicing enhances translation in mammalian cells: An additional function of the exon junction complex. Genes & Development, 18(2), 210–222.
Tan, K., Stupack, D. G., & Wilkinson, M. F. (2022). Nonsense-mediated RNA decay: an emerging modulator of malignancy. Nature Reviews Cancer, 22, 437–451.
Celik, A., Baker, R., He, F., & Jacobson, A. (2017). High-resolution profiling of NMD targets in yeast reveals translational fidelity as a basis for substrate selection. RNA, 23, 735–748.
Boehm, V., Britto-Borges, T., Steckelberg, A.-L., Singh, K. K., Gerbracht, J. V., Gueney, E., Blazquez, L., Altmüller, J., Dieterich, C., & Gehring, N. H. (2018). Exon junction complexes suppress spurious splice sites to safeguard transcriptome integrity. Molecular Cell, 72(3), 482–495.
Wang, Z., Ballut, L., Barbosa, I., & Le Hir, H. (2018). Exon Junction Complexes can have distinct functional flavours to regulate specific splicing events. Scientific Reports, 8(1), 1–8.
Aznarez, I., Nomakuchi, T. T., Tetenbaum-Novatt, J., Rahman, M. A., Fregoso, O., Rees, H., & Krainer, A. R. (2018). Mechanism of nonsense-mediated mRNA decay stimulation by splicing factor SRSF1. Cell Reports, 23(7), 2186–2198.
Rahman, M. A., Lin, K. T., Bradley, R. K., Abdel-Wahab, O., & Krainer, A. R. (2020). Recurrent SRSF2 mutations in MDS affect both splicing and NMD. Genes & Development, 34(5–6), 413–427.
Dinesh-Kumar, S. P., & Baker, B. J. (2000). Alternatively, spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proceedings of the National academy of Sciences of the United States of America, 97, 1908–1913.
Celik, A., Baker, R., He, F., & Jacobson, A. (2017). High-resolution profiling of NMD targets in yeast reveals translational fidelity as a basis for substrate selection. RNA, 23(5), 735–748.
Lynch, S. A., Nguyen, L. S., Ng, L. Y., Waldron, M., McDonald, D., & Gecz, J. (2012). Broadening the phenotype associated with mutations in UPF3B: Two further cases with renal dysplasia and variable developmental delay. European Journal of Medical Genetics, 55(8–9), 476–479.
Laumonnier, F., Shoubridge, C., Antar, C., Nguyen, L. S., Van Esch, H., Kleefstra, T., Briault, S., Fryns, J. P., Hamel, B., Chelly, J., Ropers, H. H., Ronce, N., Blesson, S., Moraine, C., Gécz, J., & Raynaud, M. (2010). Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Molecular Psychiatry, 15(7), 767–776.
Xu, X., Zhang, L., Tong, P., Xun, G., Su, W., Xiong, Z., Zhu, T., Zheng, Y., Luo, S., Pan, Y., Xia, K., & Hu, Z. (2013). Exome sequencing identifies UPF3B as the causative gene for a Chinese non-syndrome mental retardation pedigree. Clinical Genetics, 83(6), 560–564.
Jolly, L. A., Homan, C. C., Jacob, R., Barry, S., & Gecz, J. (2013). The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Human Molecular Genetics, 22(23), 4673–4687.
Alrahbeni, T., Sartor, F., Anderson, J., Miedzybrodzka, Z., McCaig, C., & Müller, B. (2015). Full UPF3B function is critical for neuronal differentiation of neural stem cells. Molecular Brain, 8(1), 1–15.
Huang, L., Shum, E. Y., Jones, S. H., Lou, C.-H., Chousal, J., Kim, H., Roberts, A. J., Jolly, L. A., Espinoza, J. L., Skarbrevik, D. M., Phan, M. H., Cook-Andersen, H., Swerdlow, N. R., Gecz, J., & Wilkinson, M. F. (2018). A Upf3b-mutant mouse model with behavioral and neurogenesis defects. Molecular Psychiatry, 23(8), 1773–1786.
Huang, L., Low, A., Damle, S., Keenan, M. M., Kuntz, S., Murray, S. F., Monia, B. P., & Guo, S. (2018). Antisense suppression of the nonsense mediated decay factor Upf3b as a potential treatment for diseases caused by nonsense mutations. Genome Biology, 19(1).
Nguyen, L. S., Jolly, L., Shoubridge, C., Chan, W. K., Huang, L., Laumonnier, F., Raynaud, M., Hackett, A., Field, M., Rodriguez, J., Srivastava, A. K., Lee, Y., Long, R., Addington, A. M., Rapoport, J. L., Suren, S., Hahn, C. N., Gamble, J., Wilkinson, M. F., … Gecz, J. (2012). Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Molecular Psychiatry, 17(11), 1103–1115.
Chan, W. K., Huang, L., Gudikote, J. P., Chang, Y. F., Imam, J. S., MacLean, J. A., & Wilkinson, M. F. (2007). An alternative branch of the nonsense-mediated decay pathway. The EMBO journal, 26(7), 1820–1830.
Kunz, J. B., Neu-Yilik, G., Hentze, M. W., Kulozik, A. E., & Gehring, N. H. (2006). Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA, 12(6), 1015–1022.
Chan, W. K., Bhalla, A. D., Le Hir, H., Nguyen, L. S., Huang, L., Gécz, J., & Wilkinson, M. F. (2009). A UPF3-mediated regulatory switch that maintains RNA surveillance. Nature Structural & Molecular Biology, 16(7), 747–753.
Avery, P., Vicente-Crespo, M., Francis, D., Nashchekina, O., Alonso, C. R., & Palacios, I. M. (2011). Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. RNA, 17(4), 624–638.
Thorén, L., Norgaard, G. A., Weischenfeldt, J., Waage, J., Jakobsen, J. S., Damgaard, I., Bergström, F., Blom, A. M., Borup, R., Bisgaard, H. C., & Porse, B. T. (2010). UPF2 is a critical regulator of liver development, function and regeneration. PLOS ONE, 5(7), e11650.
Weischenfeldt, J., Waage, J., Tian, G., Zhao, J., Damgaard, I., Jakobsen, J. S., Kristiansen, K., Krogh, A., Wang, J., & Porse, B. T. (2012). Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. GenomeBiology.com (London. Print), 13(5), R35.
Nguyen, L. S., Kim, H., Rosenfeld, J. A., Shen, Y., Gusella, J. F., Lacassie, Y., Layman, L. C., Shaffer, L. G., & Gécz, J. (2013). Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Human Molecular Genetics, 22(9), 1816–1825.
Johnson, J. L., Stoica, L. E., Liu, Y., Zhu, P., Bhattacharya, A., Buffington, S. A., Huq, R., Eissa, N. T., Larsson, O., Porse, B. T., Domingo, D., Nawaz, U., Carroll, R., Jolly, L. A., Scerri, T. S., Kim, H. G., Brignell, A., Coleman, M., Braden, R., … Costa-Mattioli, M. (2019). Inhibition of UPF2-Dependent Nonsense-Mediated decay leads to behavioral and neurophysiological abnormalities by activating the immune response. Neuron, 104(4), 665–679.e8.
Ivanov, P. V., Gehring, N. H., Kunz, J. B., Hentze, M. W., & Kulozik, A. E. (2008). Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. The EMBO Journal, 27(5), 736–747.
Karam, R., Lou, C. H., Kroeger, H., Huang, L., Lin, J. H., & Wilkinson, M. F. (2015). The unfolded protein response is shaped by the NMD pathway. EMBO Reports, 16(5), 599–609.
Lou, C.-H., Chousal, J., Goetz, A., Shum, E. Y., Brafman, D., Liao, X., Mora-Castilla, S., Ramaiah, M., Cook-Andersen, H., Laurent, L., & Wilkinson, M. F. (2016). Nonsense-mediated RNA decay influences human embryonic stem cell fate. Stem Cell Reports, 6(6), 844–857.
Gong, C., Kim, Y. K., Woeller, C. F., Tang, Y., & Maquat, L. E. (2009). SMD and NMD are competitive pathways that contribute to myogenesis: Effects on PAX3 and myogenin mRNAs. Genes & Development, 23(1), 54–66.
Gowravaram, M., Schwarz, J., Khilji, S. K., Urlaub, H., & Chakrabarti, S. (2019). Insights into the assembly and architecture of a Staufen-mediated mRNA decay (SMD)-competent mRNP. Nature Communications, 10(1), 5054.
Shum, E. Y., Jones, S. H., Shao, A., Chousal, J. N., Krause, M. D., Chan, W.-K., Lou, C.-H., Espinoza, J. L., Song, H.-W., Phan, M. H., Ramaiah, M., Huang, L., McCarrey, J. R., Peterson, K. J., De Rooij, D. G., Cook-Andersen, H., & Wilkinson, M. F. (2016). The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell, 165(2), 382–395.
Baird, T. D., Cheng, K. C. C., Chen, Y. C., Buehler, E., Martin, S. E., Inglese, J., & Hogg, J. R. (2018). ICE1 promotes the link between splicing and nonsense-mediated mRNA decay. eLife, 7, e33178.
Ryu, I., Won, Y.-S., Ha, H., Kim, E., Park, Y., Kim, M. K., Kwon, D. H., Choe, J., Song, H. K., Jung, H., & Kim, Y. K. (2019). eIF4A3 phosphorylation by CDKs affects NMD during the cell cycle. Cell Reports, 26(8), 2126–2139.
Hsu, I. W., Hsu, M., Li, C., Chuang, T. W., Lin, R. I., & Tarn, W. Y. (2005). Phosphorylation of Y14 modulates its interaction with proteins involved in mRNA metabolism and influences its methylation. Journal of Biological Chemistry, 280(41), 34507–34512.
Tatsuno, T., & Ishigaki, Y. (2018). C-terminal short arginine/serine repeat sequence-dependent regulation of Y14 (RBM8A) localization. Science and Reports, 8, 612.
Trembley, J. H., Tatsumi, S., Sakashita, E., Loyer, P., Slaughter, C. A., Suzuki, H., Endo, H., Kidd, V. J., & Mayeda, A. (2005). Activation of pre-mRNA splicing by human RNPS1 is regulated by CK2 phosphorylation. Molecular and Cellular Biology, 25(4), 1446–1457.
Viegas, M. H., Gehring, N. H., Breit, S., Hentze, M. W., & Kulozik, A. E. (2007). The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the nonsense mediated decay pathway. Nucleic Acids Research, 35(13), 4542–4551.
Bruno, I. G., Karam, R., Huang, L., Bhardwaj, A., Lou, C. H., Shum, E. Y., Song, H.-W., Corbett, M. A., Gifford, W. D., Gecz, J., Pfaff, S. L., & Wilkinson, M. F. (2011). Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Molecular Cell, 42(4), 500–510.
Baguet, A., Degot, S., Cougot, N., Bertrand, E., Chenard, M.-P., Wendling, C., Kessler, P., Hir, H. L., Rio, M.-C., & Tomasetto, C. (2007). The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. Journal of Cell Science, 120(16), 2774–2784.
Cougot, N., Daguenet, É., Baguet, A., Cavalier, A., Thomas, D., Bellaud, P., Fautrel, A., Godey, F., Bertrand, É., Tomasetto, C., & Gillet, R. (2014). MLN51 triggers P-body disassembly and formation of a new type of RNA granules. Journal of Cell Science, 127(21), 4692–4701.
Chu, V., Feng, Q., Lim, Y., & Shao, S. (2021). Selective destabilization of polypeptides synthesized from NMD-targeted transcripts. Molecular Biology of the Cell, 32, ar38.
Panigrahi, G. K., & Satapathy, K. B. (2020). Sacrificed surveillance process favours plant defense: A review. Plant Archives, 20(1), 2551–2559.
Panigrahi, G. K., Sahoo, S. K., Sahoo, A., Behera, S., Sahu, S. R., Dash, A., & Satapathy, K. B. (2021). Bioactive molecules from plants: A prospective approach to combat SARS-Cov-2. Advances in Traditional Medicine, 23, 1–14.
Kashima, I., Yamashita, A., Izumi, N., Kataoka, N., Morishita, R., Hoshino, S., Ohno, M., Dreyfuss, G., & Ohno, S. (2006). Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & Development, 20, 355–367.
Panigrahi, G. K., & Satapathy, K. B. (2021). Pseudomonas syringae pv. syringae infection orchestrates the fate of the Arabidopsis J domain containing cochaperone and decapping protein factor 5. Physiological and Molecular Plant Pathology, 113(101598), 1–9.
Sahoo, A., Satapathy, K. B., & Panigrahi, G. K. (2023). Ectopic expression of disease resistance protein promotes resistance against pathogen infection and drought stress in Arabidopsis. Physiological and Molecular Plant Pathology, 124(101949), 1–7.
Jung, H. W., Panigrahi, G. K., Jung, G.-Y., Lee, Y. J., Shin, K. H., Sahoo, A., Choi, E. S., Lee, E., Kim, K. M., Yang, S. H., Jeon, J. S., Lee, S. C., & Kim, S. H. (2020). PAMP-triggered immunity involves proteolytic degradation of core nonsense-mediated mRNA decay factors during early defense response. The Plant Cell, 32(4), 1081–1101.
Addington, A. M., Gauthier, J., Piton, A., Hamdan, F. F., Raymond, A., Gogtay, N., Miller, R., Tossell, J., Bakalar, J., & Inoff-Germain, G. (2011). A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Molecular Psychiatry, 16, 238–239.
Tan, K., Jones, S. H., Lake, B. B., Dumdie, J. N., Shum, E. Y., Zhang, L., Chen, S., Sohni, A., Pandya, S., Gallo, R. L., Zhang, K., Cook‐Andersen, H., & Wilkinson, M. (2020). The role of the NMD factor UPF3B in olfactory sensory neurons. eLife, 9, e57525.
Colak, D., Ji, S. J., Porse, B. T., & Jaffrey, S. R. (2013). Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell, 153, 1252–1265.
Bruno, I. G., Karam, R., Huang, L., Bhardwaj, A., Lou, C. H., Shum, E. Y., Song, H. W., Corbett, M. A., Gifford, W. D., & Gecz, J. (2011). Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Molecular Cell, 42, 500–510.
Agarwal, V., Bell, G. W., Nam, J. W., & Bartel, D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife, 4, e05005.
Lou, C. H., Shao, A., Shum, E. Y., Espinoza, J. L., Huang, L., Karam, R., & Wilkinson, M. F. (2014). Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Reports, 6, 748–764.
Wang, G., Jiang, B., Jia, C., Chai, B., & Liang, A. (2013). MicroRNA 125 represses nonsense-mediated mRNA decay by regulating SMG1 expression. Biochemical and Biophysical Research Communications, 435, 16–20.
Gong, C., Kim, Y. K., Woeller, C. F., Tang, Y., & Maquat, L. E. (2009). SMD and NMD are competitive pathways that contribute to myogenesis: Effects on PAX3 and myogenin mRNAs. Genes & Development, 23, 54–66.
Bourgeois, C. F., Lejeune, F., & Stévenin, J. (2004). Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Progress in Nucleic Acid Research and Molecular Biology, 78, 37–88.
Braunschweig, U., Gueroussov, S., Plocik, A. M., Graveley, B. R., & Blencowe, B. J. (2013). Dynamic integration of splicing within gene regulatory pathways. Cell, 152, 1252–1269.
Chabot, B., & Shkreta, L. (2016). Defective control of pre-messenger RNA splicing in human disease. Journal of Cell Biology, 212, 13–27.
Serin, G., Gersappe, A., Black, J. D., Aronoff, R., & Maquat, L. E. (2001). Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Molecular and Cellular Biology, 21, 209–223.
Ohnishi, T., Yamashita, A., Kashima, I., Schell, T., Anders, K. R., Grimson, A., Hachiya, T., Hentze, M. W., Anderson, P., & Ohno, S. (2003). Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Molecular Cell, 12(5), 1187–1200.
Gowravaram, M., Bonneau, F., Kanaan, J., Maciej, V. D., Fiorini, F., Raj, S., Croquette, V., Hir, H. L., & Chakrabarti, S. (2018). A conserved structural element in the RNA helicase UPF1 regulates its catalytic activity in an isoform-specific manner. Nucleic Acids Research, 46(5), 2648–2659.
Padariya, M., Fahraeus, R., Hupp, T., & Kalathiya, U. (2021). Molecular determinants and specificity of mRNA with alternatively -Spliced UPF1 isoforms, influenced by an insertion in the ‘regulatory loop.’ International Journal of Molecular Sciences, 22(23), 12744.
Longman, D., Jackson-Jones, K. A., Maslon, M. M., Murphy, L. C., Young, R. S., Stoddart, J. J., Hug, N., Taylor, M. S., Papadopoulos, D. K., & Cáceres, J. F. (2020). Identification of a localized nonsense-mediated decay pathway at the endoplasmic reticulum. Genes & Development, 34(15–16), 1075–1088.
Chiu, S. Y., Lejeune, F., Ranganathan, A. C., & Maquat, L. E. (2004). The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes & Development, 18(7), 745–754.
Nickless, A., Jackson, E., Marasa, J., Nugent, P., Mercer, R. W., Piwnica-Worms, D., & You, Z. (2014). Intracellular calcium regulates nonsense-mediated mRNA decay. Nature Medicine, 20(8), 961–966.
Tantral, L., Malathi, K., Kohyama, S., Silane, M., Berenstein, A., & Jayaraman, T. (2004). Intracellular calcium release is required for caspase-3 and-9 activation. Cell Biochemistry and Function, 22(1), 35–40.
Jia, J., Furlan, A., Gonzalez-Hilarion, S., Leroy, C., Gruenert, D. C., Tulasne, D., & Lejeune, F. (2015). Caspases shutdown nonsense-mediated mRNA decay during apoptosis. Cell Death & Differentiation, 22(11), 1754–1763.
Popp, M. W., & Maquat, L. E. (2015). Attenuation of nonsense-mediated mRNA decay facilitates the response to chemotherapeutics. Nature communications, 6(1), 6632.
Li, Z., Vuong, J. K., Zhang, M., Stork, C., & Zheng, S. (2017). Inhibition of nonsense-mediated RNA decay by ER stress. RNA, 23(3), 378–394.
Chang, L., Li, C., Guo, T., Wang, H., Ma, W., Yuan, Y., Liu, Q., Ye, Q., & Liu, Z. (2016). The human RNA surveillance factor UPF1 regulates tumorigenesis by targeting Smad7 in hepatocellular carcinoma. Journal of Experimental & Clinical Cancer Research, 35(1), 1–12.
Shi, M., Wang, S., Yao, Y., Li, Y., Zhang, H., Han, F., Nie, H., Su, J., Wang, Z., Yue, L., Cao, J., & Li, Y. (2014). Biological and clinical significance of epigenetic silencing of MARVELD1 gene in lung cancer. Scientific Reports, 4(1), 7545.
Bokhari, A., Jonchere, V., Lagrange, A., Bertrand, R., Svrcek, M., Marisa, L., Buhard, O., Greene, M., Demidova, A., Jia, J., Adriaenssens, E., Chassat, T., Biard, D. S., Flejou, J.-F., Lejeune, F., Duval, A., & Collura, A. (2018). Targeting nonsense-mediated mRNA decay in colorectal cancers with microsatellite instability. Oncogenesis, 7(9), 70.
Palma, M., Leroy, C., Salomé-Desnoulez, S., Werkmeister, E., Kong, R., Mongy, M., Hir, H. L., & Lejeune, F. (2021). A role for AKT1 in nonsense-mediated mRNA decay. Nucleic Acids Research, 49(19), 11022–11037.
Huang, L., Lou, C. H., Chan, W., Shum, E. Y., Shao, A., Stone, E., & Wilkinson, M. F. (2011). RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Molecular Cell, 43(6), 950–961.
Sahoo, A., Satapathy, K. B., & Panigrahi, G. K. (2023). Security check: plant immunity under temperature surveillance. Journal of Plant Biochemistry and Biotechnology, 33, 1–4.
Acknowledgements
Authors thank the administration and management of Centurion University of Technology and Management, Odisha, India for their heartfelt support. We apologize to all colleagues whose work could not be included owing to space limitations.
Funding
The authors would like to thank the Vice Chancellor, Centurion University of Technology and Management, Odisha for providing financial support to GKP (grant approval letter no: CUTM/VC Office/45 to GKP).
Author information
Authors and Affiliations
Contributions
All the authors have substantial contribution for the preparation of the manuscript. GKP conceptualized and conceived the idea, RD and GKP performed data curation and writing of the manuscript. All the authors have read and approved the final manuscript before submission.
Corresponding author
Ethics declarations
Competing interest
The authors declare that they have no conflicts of interest.
Ethical Approval
It is a review article. No ethics approval is required.
Consent to Publish
Not applicable.
Human and Animal Rights
It is a review article. No animals were used in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Das, R., Panigrahi, G.K. Messenger RNA Surveillance: Current Understanding, Regulatory Mechanisms, and Future Implications. Mol Biotechnol (2024). https://doi.org/10.1007/s12033-024-01062-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-024-01062-4