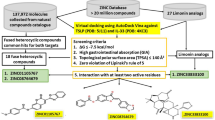
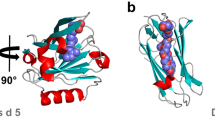
Abstract
The occurrence of allergy, a type I hypersensitivity reaction, is rising exponentially all over the world. Sometimes, allergy proves to be fatal for atopic patients, due to the occurrence of anaphylaxis. This study is aimed to find an anti-allergic agent that can inhibit the binding of IgE to Human High Affinity IgE Receptor (FCεRI), thereby preventing the degranulation of mast cells. A considerable number of potential anti-allergic compounds were assessed for their inhibitory strength through ADMET studies. AUTODOCK was used for estimating the binding energy between anti-allergic compounds and FCεRI, along with the interacting amino acids. The docked pose showing favorable binding energy was subjected to molecular dynamics simulation study. Marrubiin, a diterpenoid lactone from Lamiaceae, and epicatechin-3-gallate appears to be effective in blocking the Human High Affinity IgE Receptor (FCεRI). This in-silico study proposes the use of marrubiin and epicatechin-3-gallate, in the downregulation of allergic responses. Due to the better inhibition constant, future direction of this study is to analyze the safety and efficacy of marrubiin in anti-allergic activities through in-vivo clinical human trials.



Similar content being viewed by others
Data Availability
The PDB files of FcεRI [PDB ID: 1F2Q] is available in the Protein Data Bank (https://www.rcsb.org/). The PDB file of the ligand marrubiin is available in PubChem (https://pubchem.ncbi.nlm.nih.gov/).
Code Availability
Not Applicable.
Abbreviations
- Ag:
-
Antigen
- FcεRI:
-
High affinity IgE receptor
- IP3:
-
Inositol-1,4,5-trisphosphate
- ITAM:
-
Immunoreceptor tyrosine-based activation motif
- MC:
-
Mast cell
- PAINS:
-
Pan assay interference compounds
- PI3K:
-
Phospho-inositide 3-kinase
- PKC:
-
Protein kinase C
- RMSD:
-
Root means square deviation
References
Nagata, Y., & Suzuki, R. (2022). FcεRI: A master regulator of mast cell functions. Cells, 11(4), 622. https://doi.org/10.3390/cells11040622
Dispenza, M. C., Bochner, B. S., & MacGlashan, D. W., Jr. (2020). Targeting the FcεRI pathway as a potential strategy to prevent food-induced anaphylaxis. Frontiers in Immunology, 11, 614402. https://doi.org/10.3389/fimmu.2020.614402
Ando, T., & Kitaura, J. (2021). Tuning IgE: IgE-associating molecules and their effects on IgE-dependent mast cell reactions. Cells, 10(7), 1697. https://doi.org/10.3390/cells10071697
Lecce, M., Molfetta, R., Milito, N. D., Santoni, A., & Paolini, R. (2020). FcεRI Signaling in the modulation of allergic response: Role of mast cell-derived exosomes. International Journal of Molecular Sciences, 21(15), 5464. https://doi.org/10.3390/ijms21155464
Sakai, S., Sugawara, T., Matsubara, K., & Hirata, T. (2009). Inhibitory effect of carotenoids on the degranulation of mast cells via suppression of antigen-induced aggregation of high affinity IgE receptors. The Journal of Biological Chemistry, 284(41), 28172–28179. https://doi.org/10.1074/jbc.M109.001099
Tokura, T., Nakano, N., Ito, T., Matsuda, H., Nagasako-Akazome, Y., Kanda, T., Ikeda, M., Okumura, K., Ogawa, H., & Nishiyama, C. (2005). Inhibitory effect of polyphenol-enriched apple extracts on mast cell degranulation in vitro targeting the binding between IgE and FcepsilonRI. Bioscience, Biotechnology, and Biochemistry, 69(10), 1974–1977. https://doi.org/10.1271/bbb.69.1974
Vo, T. S. (2020). Natural products targeting FcεRI receptor for anti-allergic therapeutics. Journal of Food Biochemistry, 44(8), 13335. https://doi.org/10.1111/jfbc.13335
Popoola, O. K., Elbagory, A. M., Ameer, F., & Hussein, A. A. (2013). Marrubiin. Molecules (Basel, Switzerland), 18(8), 9049–9060. https://doi.org/10.3390/molecules18089049
Sim, L. Y., Abd Rani, N. Z., & Husain, K. (2019). Lamiaceae: An insight on their anti-allergic potential and its mechanisms of action. Frontiers in Pharmacology, 10, 677. https://doi.org/10.3389/fphar.2019.00677
Laha, A., Bandopadhyay, R., Sarkar, A., Chakraborty, P., & Panja, A. S. (2023). Efficacy screening of prospective anti-allergic drug candidates: An in silico study. Current Bioinformatics, 18(2), 143–153. https://doi.org/10.2174/1574893618666221019092212
Mnonopi, N., Levendal, R. A., Davies-Coleman, M. T., & Frost, C. L. (2011). The cardioprotective effects of marrubiin, a diterpenoid found in Leonotis leonurus extracts. Journal of Ethnopharmacology, 138(1), 67–75. https://doi.org/10.1016/j.jep.2011.08.041
Aćimović, M., Jeremić, K., Salaj, N., Gavarić, N., Kiprovski, B., Sikora, V., & Zeremski, T. (2020). Marrubium vulgare L.: A phytochemical and pharmacological overview. Molecules (Basel, Switzerland), 25(12), 2898. https://doi.org/10.3390/molecules25122898
Kapoor, Y., & Kumar, K. (2020). Structural and clinical impact of anti-allergy agents: An overview. Bioorganic Chemistry, 94, 103351. https://doi.org/10.1016/j.bioorg.2019.103351
Burley, S. K., Bhikadiya, C., Bi, C., Bittrich, S., Chen, L., Crichlow, G. V., Duarte, J. M., Dutta, S., Fayazi, M., Feng, Z., Flatt, J. W., Ganesan, S. J., Goodsell, D. S., Ghosh, S., Kramer Green, R., Guranovic, V., Henry, J., Hudson, B. P., Lawson, C. L., … Zardecki, C. (2022). RCSB Protein Data Bank: Celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3D. Protein Science, 31(1), 187–208. https://doi.org/10.1002/pro.4213
Kim, S., Cheng, T., He, S., Thiessen, P. A., Li, Q., Gindulyte, A., & Bolton, E. E. (2022). PubChem protein, gene, pathway, and taxonomy data collections: bridging biology and chemistry through target-centric views of PubChem data. Journal of molecular biology,434(11), 167514.https://doi.org/10.1016/j.jmb.2022.167514
Xiong, G., Wu, Z., Yi, J., Fu, L., Yang, Z., Hsieh, C., Yin, M., Zeng, X., Wu, C., Lu, A., Chen, X., Hou, T., & Cao, D. (2021). ADMETlab 20: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Research, 49(W1), W5–W14. https://doi.org/10.1093/nar/gkab255
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. https://doi.org/10.1002/jcc.21256
Ahmad, M. I., Potshangbam, A. M., Javed, M., & Ahmad, M. (2020). Studies on conformational changes induced by binding of pendimethalin with human serum albumin. Chemosphere, 243, 125270. https://doi.org/10.1016/j.chemosphere.2019.125270
Bjelkmar, P., Larsson, P., Cuendet, M. A., Hess, B., & Lindahl, E. (2010). Implementation of the CHARMM force field in GROMACS: Analysis of protein stability effects from correction maps, virtual interaction sites, and water models. Journal of Chemical Theory and Computation, 6(2), 459–466. https://doi.org/10.1021/ct900549r
GROMACS development team. (2021). Gromacs Documentation Release 2021.2. https://doi.org/10.5281/zenodo.4723561
Grace Development Team. (2008). Grace. https://plasma-gate.weizmann.ac.il/Grace/
Attique, S. A., Hassan, M., Usman, M., Atif, R. M., Mahboob, S., Al-Ghanim, K. A., Bilal, M., & Nawaz, M. Z. (2019). A molecular docking approach to evaluate the pharmacological properties of natural and synthetic treatment candidates for use against hypertension. International Journal of Environmental Research and Public Health, 16(6), 923. https://doi.org/10.3390/ijerph16060923
Abdullahi, M., & Adeniji, S. E. (2020). In-silico molecular docking and ADME/pharmacokinetic prediction studies of some novel carboxamide derivatives as anti-tubercular agents. Chemistry Africa, 3, 989–1000. https://doi.org/10.1007/s42250-020-00162-3
Umar, A. B., Uzairu, A., Shallangwa, G. A., & Uba, S. (2020). Design of potential anti-melanoma agents against SK-MEL-5 cell line using QSAR modeling and molecular docking methods. SN Applied Sciences., 2, 815. https://doi.org/10.1007/s42452-020-2620-8
Lipinski, C. A. (2004). Lead- and drug-like compounds: The rule-of-five revolution. Drug Discovery Today, 1(4), 337–341. https://doi.org/10.1016/j.ddtec.2004.11.007
Chagas, C. M., Moss, S., & Alisaraie, L. (2018). Drug metabolites and their effects on the development of adverse reactions: Revisiting Lipinski’s Rule of Five. International journal of pharmaceutics, 549(1–2), 133–149. https://doi.org/10.1016/j.ijpharm.2018.07.046
Volpe, D. A. (2011). Drug-permeability and transporter assays in Caco-2 and MDCK cell lines. Future Medicinal Chemistry, 3(16), 2063–2077. https://doi.org/10.4155/fmc.11.149
McGill, M. R., & Jaeschke, H. (2019). Biomarkers of drug-induced liver injury. Advances in Pharmacology (San Diego, Calif), 85, 221–239. https://doi.org/10.1016/bs.apha.2019.02.001
Wang, J., & Hou, T. (2015). Advances in computationally modeling human oral bioavailability. Advanced Drug Delivery Reviews, 86, 11–16. https://doi.org/10.1016/j.addr.2015.01.001
Holt, K., Nagar, S., & Korzekwa, K. (2019). Methods to predict volume of distribution. Current Pharmacology Reports, 5(5), 391–399. https://doi.org/10.1007/s40495-019-00186-5
Svennebring, A. (2016). The connection between plasma protein binding and acute toxicity as determined by the LD50 value. Drug Development Research, 77(1), 3–11. https://doi.org/10.1002/ddr.21286
McCormick, A., Swaisland, H., Reddy, V. P., Learoyd, M., & Scarfe, G. (2018). In vitro evaluation of the inhibition and induction potential of olaparib, a potent poly(ADP-ribose) polymerase inhibitor, on cytochrome P450. Xenobiotica, 48(6), 555–564. https://doi.org/10.1080/00498254.2017.1346332
Smith, D. A., Beaumont, K., Maurer, T. S., & Di, L. (2018). Relevance of half-life in drug design. Journal of Medicinal Chemistry, 61(10), 4273–4282. https://doi.org/10.1021/acs.jmedchem.7b00969
Wempe, M. F. (2022). New insights into ion channels: Predicting hERG-drug interactions. International Journal of Molecular Sciences, 23(18), 10732. https://doi.org/10.3390/ijms231810732
Zeiger, E. (2019). The test that changed the world: The Ames test and the regulation of chemicals. Mutation Research Genetic Toxicology and Environmental Mutagenesis, 841, 43–48. https://doi.org/10.1016/j.mrgentox.2019.05.007
Sepay, N., Sekar, A., Halder, U. C., Alarifi, A., & Afzal, M. (2021). Anti-COVID-19 terpenoid from marine sources: A docking, admet and molecular dynamics study. Journal of Molecular Structure, 1228, 129433. https://doi.org/10.1016/j.molstruc.2020.129433
Papadatos, G., Davies, M., Dedman, N., Chambers, J., Gaulton, A., Siddle, J., Koks, R., Irvine, S. A., Pettersson, J., Goncharoff, N., Hersey, A., & Overington, J. P. (2016). SureChEMBL: A large-scale, chemically annotated patent document database. Nucleic acids research, 44(D1), D1220–D1228. https://doi.org/10.1093/nar/gkv1253
Yang, Z. Y., Yang, Z. J., Lu, A. P., Hou, T. J., & Cao, D. S. (2021). Scopy: an integrated negative design python library for desirable HTS/VS database design. Briefings in Bioinformatics, 22(3), 194. https://doi.org/10.1093/bib/bbaa194
Baell, J. B., & Holloway, G. A. (2010). New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. Journal of Medicinal Chemistry, 53(7), 2719–2740. https://doi.org/10.1021/jm901137j
Bickerton, G. R., Paolini, G. V., Besnard, J., Muresan, S., & Hopkins, A. L. (2012). Quantifying the chemical beauty of drugs. Nature chemistry, 4(2), 90–98. https://doi.org/10.1038/nchem.1243
Dhorajiwala, T., Halder, S., & Samant, L. (2019). Comparative in silico molecular docking analysis of L-threonine-3-dehydrogenase, a protein target against african trypanosomiasis using selected phytochemicals. Journal of Applied Biotechnology Reports, 6(3), 101–108. https://doi.org/10.29252/JABR.06.03.04
Garman, S. C., Kinet, J. P., & Jardetzky, T. S. (1998). Crystal structure of the human high-affinity IgE receptor. Cell, 95(7), 951–961. https://doi.org/10.1016/s0092-8674(00)81719-5
Laha, A., Bandopadhyay, R., & Panja, A. S. (2021). Structural phylogeny of different allergens may reveal common epitopic footprint. Protein and Peptide Letters, 28(10), 1099–1107. https://doi.org/10.2174/0929866528666210622145710
Acknowledgements
We are thankful to UGC-Center of Advanced Study, Department of Botany, The University of Burdwan, Burdwan and DST-FIST [DST No. SR/FST/LS-1/2018/188(C)] for providing us with all the necessary research facilities.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
RB conceptualized the project. AL performed the in-silico study under the supervision of RB and ASP. AL, AS and ASP have analyzed the data. The analysis tools were provided by ASP and RB. AL wrote the manuscript. AS helped in writing the draft paper. All authors have edited and read the manuscript.
Corresponding author
Ethics declarations
Competing interest
Anubhab Laha, Aniket Sarkar, Anindya Sundar Panja and Rajib Bandopadhyay declare that this article content has no conflict of interest.
Consent to Participate
This article contains information and analysis of in-silico data only, and it does not involve studies with animal or human subjects performed by any of the authors. Thus the need for consent to participate is not applicable for this research article.
Consent for Publication
This article does not contain studies with animal or human subjects performed by any of the authors. So, requirement of consent for publication is inapplicable for this article.
Ethical Approval
This article contains information and analysis of in-silico data only. This article does not involve studies with animal or human subjects performed by any of the authors, which should be approved by Ethics Committee.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Laha, A., Sarkar, A., Panja, A.S. et al. Screening of Prospective Antiallergic Compound as FcεRI Inhibitors and Its Antiallergic Efficacy Through Immunoinformatics Approaches. Mol Biotechnol 66, 26–33 (2024). https://doi.org/10.1007/s12033-023-00728-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-023-00728-9