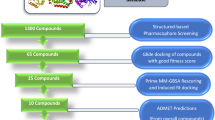
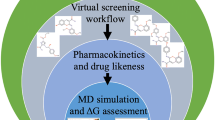
Abstract
Implementation of computational tools in the identification of novel drug targets for Tuberculosis (TB) has been a promising area of research. TB has been a chronic infectious disease caused by Mycobacterium tuberculosis (Mtb) localized primarily on the lungs and it has been one of the most successful pathogen in the history of mankind. Extensively arising drug resistivity in TB has made it a global challenge and need for new drugs has become utmost important.The involvement of Nucleoid-Associated Proteins (NAPs) in maintaining the structure of the genomic material and regulating various cellular processes like transcription, DNA replication, repair and recombination makes significant, has opened a new arena to find the drugs targeting Mtb. The current study aims to identify potential inhibitors of NAPs through a computational approach. In the present work we worked on the eight NAPs of Mtb, namely, Lsr2, EspR, HupB, HNS, NapA, mIHF and NapM. The structural modelling and analysis of these NAPs were carried out. Moreover, molecular interaction were checked and binding energy was identified for 2500 FDA-approved drugs that were selected for antagonist analysis to choose novel inhibitors targeting NAPs of Mtb. Drugs including Amikacin, streptomycin, kanamycin, and isoniazid along with eight FDA-approved molecules that were found to be potential novel targets for these mycobacterial NAPs and have an impact on their functions. The potentiality of several anti-tubercular drugs as therapeutic agents identified through computational modelling and simulation unlocks a new gateway for accomplishing the goal to treat TB.
Graphical Abstract
Complete framework of the methodology employed in this study to predict inhibitors against mycobacterial NAPs.




Similar content being viewed by others
Data Availability
All data generated or analysed during this study are included in this published article. The Data used in the study was provided by DrugBank (on request).
References
Singhvi, N., Gupta, V., Gaur, M., Sharma, V., Puri, A., Singh, Y., Dubey, G. P., & Lal, R. (2020). Interplay of human gut microbiome in health and wellness. Indian Journal of Microbiology, 60(1), 26–36.
Mohammadzadeh, R., Ghazvini, K., Farsiani, H., & Soleimanpour, S. (2021). Mycobacterium tuberculosis extracellular vesicles: Exploitation for vaccine technology and diagnostic methods. Critical Reviews in Microbiology, 47(1), 13–33.
Bhargava, A., Bhargava, M., & Juneja, A. (2021). Social determinants of tuberculosis: context, framework, and the way forward to ending TB in India. Expert Review of Respiratory Medicine, 15(7), 867–883.
Sudbury, E. L., Clifford, V., Messina, N. L., Song, R., & Curtis, N. (2020). Mycobacterium tuberculosis-specific cytokine biomarkers to differentiate active TB and LTBI: a systematic review. The Journal of infection, 81(6), 873–881.
Rowneki, M., Aronson, N., Du, P., Sachs, P., Blakemore, R., Chakravorty, S., Levy, S., Jones, A. L., Trivedi, G., Chebore, S., Addo, D., Byarugaba, D. K., Njobvu, P. D., Wabwire-Mangen, F., Erima, B., Ramos, E. S., Evans, C. A., Hale, B., Mancuso, J. D., & Alland, D. (2020). Detection of drug resistant mycobacterium tuberculosis by high-throughput sequencing of DNA isolated from acid fast bacilli smears. PLoS ONE, 15(5), e0232343.
Ding, C., Wang, S., Shangguan, Y., Feng, X., Guo, W., Shi, P., Ji, Z., & Xu, K. (2020). Epidemic trends of tuberculosis in China from 1990 to 2017: Evidence from the global burden of disease study. Infection and Drug Resistance, 13, 1663–1672.
Seddon, J. A., Johnson, S., Palmer, M., van der Zalm, M. M., Lopez-Varela, E., Hughes, J., & Schaaf, H. S. (2021). Multidrug-resistant tuberculosis in children and adolescents: Current strategies for prevention and treatment. Expert Review of Respiratory Medicine, 15(2), 221–237.
Migliori, G. B., Tiberi, S., Zumla, A., Petersen, E., Chakaya, J. M., Wejse, C., & Zellweger, J. P. (2020). MDR/XDR-TB management of patients and contacts: challenges facing the new decade. The 2020 clinical update by the global tuberculosis network. International journal of infectious diseases IJID Official Publication of the International Society for Infectious Diseases, 92S, S15–S25.
Silva, J. V., Santos, S., Machini, M. T., & Giarolla, J. (2021). Neglected tropical diseases and infectious illnesses: Potential targeted peptides employed as hits compounds in drug design. Journal of Drug Targeting, 29(3), 269–283.
Singh, Y., Beamer, G., Sun, X., & Shukla, P. (2022). Recent developments in systems biology and genetic engineering toward design of vaccines for TB. Critical Reviews in Biotechnology, 42(4), 532–547.
Stojkova, P., Spidlova, P., & Stulik, J. (2019). Nucleoid-associated protein HU: A lilliputian in gene regulation of bacterial virulence. Frontiers in Cellular and Infection Microbiology, 9, 159.
Swiercz, J. P., Nanji, T., Gloyd, M., Guarné, A., & Elliot, M. A. (2013). A novel nucleoid-associated protein specific to the actinobacteria. Nucleic Acids Research, 41(7), 4171–4184.
Odermatt, N. T., Sala, C., Benjak, A., & Cole, S. T. (2018). Essential nucleoid associated protein mIHF (Rv1388) controls virulence and housekeeping genes in mycobacterium tuberculosis. Scientific Reports, 8(1), 14214.
Basu, D., Khare, G., Singh, S., Tyagi, A., Khosla, S., & Mande, S. C. (2009). A novel nucleoid-associated protein of mycobacterium tuberculosis is a sequence homolog of GroEL. Nucleic Acids Research, 37(15), 4944–4954.
Kriel, N. L., Gallant, J., van Wyk, N., van Helden, P., Sampson, S. L., Warren, R. M., & Williams, M. J. (2018). Mycobacterial nucleoid associated proteins: An added dimension in gene regulation. Tuberculosis (Edinburgh, Scotland), 108, 169–177.
Sritharan, M. (2016). Iron homeostasis in mycobacterium tuberculosis: Mechanistic insights into siderophore-mediated iron uptake. Journal of bacteriology, 198(18), 2399–2409.
Hołówka, J., Trojanowski, D., Janczak, M., Jakimowicz, D., & Zakrzewska-Czerwińska, J. (2018). The origin of chromosomal replication is asymmetrically positioned on the mycobacterial nucleoid, and the timing of its firing depends on HupB. Journal of Bacteriology, 200(10), e00044-e118.
Pule, C. M., Sampson, S. L., Warren, R. M., Black, P. A., van Helden, P. D., Victor, T. C., & Louw, G. E. (2016). Efflux pump inhibitors: Targeting mycobacterial efflux systems to enhance TB therapy. The Journal of Antimicrobial Chemotherapy, 71(1), 17–26.
Chaudhari, K., Surana, S., Jain, P., & Patel, H. M. (2016). Mycobacterium tuberculosis (MTB) GyrB inhibitors: An attractive approach for developing novel drugs against TB. European Journal of Medicinal Chemistry, 124, 160–185.
Sabe, V. T., Tolufashe, G. F., Ibeji, C. U., Maseko, S. B., Govender, T., Maguire, G., Lamichhane, G., Honarparvar, B., & Kruger, H. G. (2019). Identification of potent L, D-transpeptidase 5 inhibitors for Mycobacterium tuberculosis as potential anti-TB leads: Virtual screening and molecular dynamics simulations. Journal of Molecular Modeling, 25(11), 328.
Dame, R. T., Rashid, F. M., & Grainger, D. C. (2020). Chromosome organization in bacteria: Mechanistic insights into genome structure and function. Nature Reviews Genetics, 21(4), 227–242.
Libby, P., Ridker, P. M., & Hansson, G. K. (2011). Progress and challenges in translating the biology of atherosclerosis. Nature, 473(7347), 317–325.
Wang, J., Wu, M. Y., Tan, J. Q., Li, M., & Lu, J. H. (2019). High content screening for drug discovery from traditional Chinese medicine. Chinese Medicine, 14, 5.
Tong, J. C., & Ren, E. C. (2009). Immunoinformatics: Current trends and future directions. Drug Discovery Today, 14(13–14), 684–689.
Hasan, M., Islam, S., Chakraborty, S., Mustafa, A. H., Azim, K. F., Joy, Z. F., Hossain, M. N., Foysal, S. H., & Hasan, M. N. (2020). Contriving a chimeric polyvalent vaccine to prevent infections caused by herpes simplex virus (type-1 and type-2): An exploratory immunoinformatic approach. Journal of Biomolecular Structure & Dynamics, 38(10), 2898–2915.
Zheng, M., Zhao, J., Cui, C., Fu, Z., Li, X., Liu, X., Ding, X., Tan, X., Li, F., Luo, X., Chen, K., & Jiang, H. (2018). Computational chemical biology and drug design: Facilitating protein structure, function, and modulation studies. Medicinal Research Reviews, 38(3), 914–950.
Sunita, S., & A., Singh, Y., & Shukla, P. (2020). Computational tools for modern vaccine development. Human Vaccines & Immunotherapeutics, 16(3), 723–735.
Yuan, P., He, L., Chen, D., Sun, Y., Ge, Z., Shen, D., & Lu, Y. (2020). Proteomic characterization of Mycobacterium tuberculosis reveals potential targets of bostrycin. Journal of Proteomics, 212, 103576.
Schwede, T., Kopp, J., Guex, N., & Peitsch, M. C. (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Research, 31(13), 3381–3385.
Guex, N., Peitsch, M. C., & Schwede, T. (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis, 30(Suppl 1), S162–S173.
Muehlemann, S. (2016). Making apprenticeships profitable for firms and apprentices: The Swiss model. Challenge, 59(5), 390–404.
Goddard, T. D., Brilliant, A. A., Skillman, T. L., Vergenz, S., Tyrwhitt-Drake, J., Meng, E. C., & Ferrin, T. E. (2018). Molecular visualization on the holodeck. Journal of Molecular Biology, 430(21), 3982–3996.
Wiederstein, M., & Sippl, M. J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research, 35(2), W407–W410.
Cristobal, S., Zemla, A., Fischer, D., Rychlewski, L., & Elofsson, A. (2001). A study of quality measures for protein threading models. BMC Bioinformatics, 2(1), 5.
Burley, S. K., Berman, H. M., Bhikadiya, C., Bi, C., Chen, L., Di Costanzo, L., Christie, C., Dalenberg, K., Duarte, J. M., Dutta, S., & Feng, Z. (2019). RCSB protein data bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Research, 47(D1), D464–D474.
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., Sajed, T., Johnson, D., Li, C., Sayeeda, Z., Assempour, N., Iynkkaran, I., Liu, Y., Maciejewski, A., Gale, N., Wilson, A., Chin, L., Cummings, R., Le, D., Pon, A., Knox, C., & Wilson, M. (2017). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Research, 46(D1), D1074–D1082. https://doi.org/10.1093/nar/gkx1037
O’Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open babel: An open chemical toolbox. Journal of Cheminformatics, 3(1), 33.
Tian, W., Chen, C., Lei, X., Zhao, J., & Liang, J. (2018). CASTp 30: Computed atlas of surface topography of proteins. Nucleic Acids Research, 46(1), W363–W367.
Rajasekhar, S., Karuppasamy, R., & Chanda, K. (2021). Exploration of potential inhibitors for tuberculosis via structure-based drug design, molecular docking, and molecular dynamics simulation studies. Journal of Computational Chemistry, 42(24), 1736–1749.
Tanwar, G., & Purohit, R. (2019). Gain of native conformation of Aurora A S155R mutant by small molecules. Journal of Cellular Biochemistry, 120(7), 11104–11114.
Rosário-Ferreira, N., Baptista, S. J., Barreto, C. A., Rodrigues, F. E., Silva, T. F., Ferreira, S. G., & Moreira, I. S. (2021). In silico end-to-end protein-ligand interaction characterization pipeline: The case of SARS-CoV-2. ACS Synthetic Biology, 10(11), 3209–3235.
Mousquer, G. T., Peres, A., & Fiegenbaum, M. (2020). Pathology of TB/COVID-19 co-infection: The phantom menace. Tuberculosis, 126, 102020.
Singh, P. K., Joseph, J., Goyal, S., Grover, A., & Shukla, P. (2016). Functional analysis of the binding model of microbial inulinases using docking and molecular dynamics simulation. Journal of Molecular Modeling, 22(4), 69.
Uddin, R., Siddiqui, Q. N., Azam, S. S., Saima, B., & Wadood, A. (2018). Identification and characterization of potential druggable targets among hypothetical proteins of extensively drug resistant Mycobacterium tuberculosis (XDR KZN 605) through subtractive genomics approach. European Journal of Pharmaceutical Sciences, 114, 13–23.
Yang, Z., Zeng, X., & Tsui, S. K. W. (2019). Investigating function roles of hypothetical proteins encoded by the Mycobacterium tuberculosis H37Rv genome. BMC Genomics, 20(1), 394.
Chan, H. S., Shan, H., Dahoun, T., Vogel, H., & Yuan, S. (2019). Advancing drug discovery via artificial intelligence. Trends in Pharmacological Sciences, 40(8), 592–604.
Lipinski, C., Maltarollo, V., Oliveira, P., da Silva, A., & Honorio, K. (2019). Advances and perspectives in applying deep learning for drug design and discovery. Frontiers in Robotics and AI, 6, 108.
Datta, C., Jha, R. K., Ganguly, S., & Nagaraja, V. (2019). NapA (Rv0430), a novel nucleoid-associated protein that regulates a virulence operon in Mycobacterium tuberculosis in a supercoiling-dependent manner. Journal of Molecular Biology, 431(8), 1576–1591.
Blasco, B., Chen, J. M., Hartkoorn, R., Sala, C., Uplekar, S., Rougemont, J., & …& Cole, S. T. (2012). Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathogens, 8(3), e1002621.
Jin, C., Wu, X., Dong, C., Li, F., Fan, L., Xiong, S., & Dong, Y. (2019). EspR promotes mycobacteria survival in macrophages by inhibiting MyD88 mediated inflammation and apoptosis. Tuberculosis, 116, 22–31.
Sui, J., Qiao, W., Xiang, X., & Luo, Y. (2022). Epigenetic changes in Mycobacterium tuberculosis and its host provide potential targets or biomarkers for drug discovery and clinical diagnosis. Pharmacological Research, 179, 106195.
Liu, Y., Xie, Z., Zhou, X., Li, W., Zhang, H., & He, Z. G. (2019). NapM enhances the survival of Mycobacterium tuberculosis under stress and in macrophages. Communications Biology, 2(1), 1–9.
Pinault, L., Han, J. S., Kang, C. M., Franco, J., & Ronning, D. R. (2013). Zafirlukast inhibits complexation of Lsr2 with DNA and growth of Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy, 57(5), 2134–2140.
Odermatt, N. T., Sala, C., Benjak, A., Kolly, G. S., Vocat, A., Lupien, A., & Cole, S. T. (2017). Rv3852 (H-NS) of Mycobacterium tuberculosis is not involved in nucleoid compaction and virulence regulation. Journal of Bacteriology. https://doi.org/10.1128/JB.00129-17
Shi, W., Zhang, S., Feng, J., Cui, P., Zhang, W., & Zhang, Y. (2017). Clofazimine targets essential nucleoid associated protein, mycobacterial integration host factor (mIHF), in Mycobacterium tuberculosis. bioRxiv. https://doi.org/10.1101/192161
Maji, A., Misra, R., Dhakan, D. B., Gupta, V., Mahato, N. K., Saxena, R., Mittal, P., Thukral, N., Sharma, E., Singh, A., Virmani, R., Gaur, M., Singh, H., Hasija, Y., Arora, G., Agrawal, A., Chaudhry, A., Khurana, J. P., Sharma, V. K., … Singh, Y. (2018). Gut microbiome contributes to impairment of immunity in pulmonary tuberculosis patients by alteration of butyrate and propionate producers. Environmental Microbiology, 20(1), 402–419.
Singhvi, N., Singh, P., Prakash, O., Gupta, V., Lal, S., Bechthold, A., Singh, Y., Singh, R. K., & Lal, R. (2021). Differential mass spectrometry-based proteome analyses unveil major regulatory hubs in rifamycin B production in Amycolatopsis mediterranei. Journal of Proteomics, 239, 104168.
Acknowledgements
PS acknowledges, the Lab Infrastructure grant by BHU, Varanasi (F(C)/XVIII-Spl.Fund/Misc/Infrastructure/Instt.Sc/2019–2020/10290) and BTiS-NET-Sub-Distributed Information Centre, funded by DBT, Govt. of India at the School of Biotechnology, Banaras Hindu University, Varanasi, India. Authors acknowledge DrugBank for providing the FDA approved molecules used in this study. PS also acknowledges ‘Faculty Incentive Grant’ by Institute of Eminence (IoE) Scheme by BHU, Varanasi (Letter No R/ Dev/D/IoE/Seed & Incentive/2022-23/50024).
Author information
Authors and Affiliations
Contributions
S: methodology, writing- original draft preparation. NS: software validation, Data curation, visualization, writing- original draft preparation. VG: Software validation, Data curation, Visualization. YS: writing- reviewing and editing and supervision. PS: conceptualization, writing- reviewing and editing and supervision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sunita, Singhvi, N., Gupta, V. et al. Computational Approaches for the Structure-Based Identification of Novel Inhibitors Targeting Nucleoid-Associated Proteins in Mycobacterium Tuberculosis. Mol Biotechnol 66, 814–823 (2024). https://doi.org/10.1007/s12033-023-00710-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-023-00710-5