Abstract
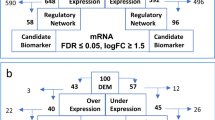
Today, Monoclonal Gammopathy of Undetermined Significance (MGUS) is known as a plasma cell malignancy susceptible to evolving into the life-threatening stage, multiple myeloma (MM), without prominent clinical manifestations. Despite the discovery of advanced therapies and multiple pathogenic markers, the complexity of MM development has made it an incurable malignancy. In this study, the microarray dataset was downloaded from the Gene Expression Omnibus (GEO) database and analyzed using the LIMMA package of R-software to determine differentially expressed genes (DEGs) in MGUS and MM compared to the control samples. Enrichment analysis of DEGs was evaluated using the GeneCodis4 software. Protein–protein interaction (PPI) networks were constructed via the GeneMANIA database, and Cytoscape visualized them. The Molecular Complex Detection (MCODE) plugin from Cytoscape was used to identify the key modules from the PPI network. Afterward, the hub genes were recognized using the cytoHubba plug-in in Cytoscape. Eventually, the correlation between hub-DEGs and MM-specific survival was evaluated via the PrognoScan database. A total of 138 (MM-normal) and 136 (MGUS-normal) DEGs were obtained from the datasets, and 62 common DEGs between MGUS and MM diseases (26 up-regulated and 36 down-regulated genes) were screened out for subsequent analyses. Following enrichment analyses and the PPI network's evaluation, FOS, FOSB, JUN, MAFF, and PPP1R15A involved in the progression of MGUS to MM were detected as the hub genes. The survival analysis revealed that FOS, FOSB, and JUN among hub genes were significantly associated with disease-specific survival (DSS) in MM. Identifying the genes involved in the progression of MGUS to MM can help in the design of preventive strategies as well as the treatment of patients. In addition, their evaluation can be effective in the survival of patients.







Similar content being viewed by others
Data Availability
This is a review study, and it is not an original. Data availability is corresponding author responsibility.
References
Lomas, O. C., Mouhieddine, T. H., Tahri, S., & Ghobrial, I. M. (2020). Monoclonal gammopathy of undetermined significance (MGUS)—Not so asymptomatic after all. Cancers, 12(6), 1554.
Jin, K., Yan, Y., Chen, M., Wang, J., Pan, X., Liu, X., et al. (2022). Multimodal deep learning with feature level fusion for identification of choroidal neovascularization activity in age-related macular degeneration. Acta Ophthalmologica, 100(2), e512–e520.
Korde, N., Kristinsson, S. Y., & Landgren, O. (2011). Monoclonal gammopathy of undetermined significance (MGUS) and smoldering multiple myeloma (SMM): Novel biological insights and development of early treatment strategies. Blood: The Journal of the American Society of Hematology, 117(21), 5573–5581.
Ng, Y.-F., Chionh, C.-Y., Torres De Guzman, M. R., Nagarajan, C., & Loh, H.-L. (2021). Lambda light chain crystalline proximal tubulopathy with probable light chain cast nephropathy and clonal plasma cell infiltrate—Uncommon manifestations of a rare form of multiple myeloma. Journal of Nephropathology. https://doi.org/10.34172/jnp.2021.08
Yan, J., Yao, Y., Yan, S., Gao, R., Lu, W., & He, W. (2020). Chiral protein supraparticles for tumor suppression and synergistic immunotherapy: An enabling strategy for bioactive supramolecular chirality construction. Nano Letters, 20(8), 5844–5852.
Pang, L., Rajkumar, S. V., Kapoor, P., Buadi, F., Dispenzieri, A., Gertz, M., et al. (2021). Prognosis of young patients with monoclonal gammopathy of undetermined significance (MGUS). Blood Cancer Journal, 11(2), 1–8.
Sekkouri, K. A., Batta, F. Z., Belghiti, K. A., Alaoui, H., Bahae, H. S., Arrayhani, M., et al. (2016). Non-amyloid deposits glomerulopathy with multiple myeloma; a rare presentation. Immunopathologia Persa, 2(1), e08.
Cancarini, G., Terlizzi, V., Garatti, A., Zeni, L., Tonoli, M., Pezzini, E., et al. (2021). Supportive treatment for cast nephropathy in patients with multiple myeloma; a pilot study. Journal of Nephropharmacology, 10(2), e20.
Landgren, O., Kyle, R. A., Pfeiffer, R. M., Katzmann, J. A., Caporaso, N. E., Hayes, R. B., et al. (2009). Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: A prospective study. Blood: The Journal of the American Society of Hematology, 113(22), 5412–5417.
Zhuo, Z., Wan, Y., Guan, D., Ni, S., Wang, L., Zhang, Z., et al. (2020). A loop-based and AGO-Incorporated virtual screening model targeting AGO-Mediated miRNA–mRNA interactions for drug discovery to rescue bone phenotype in genetically modified Mice. Advanced Science, 7(13), 1903451.
Choi, Y., Wang, J., Zhu, Y., & Lai, W.-F. (2021). Student’s perception and expectation towards pharmacy education: A qualitative study of pharmacy students in a developing country. Indian Journal of Pharmaceutical Education and Research, 55(1), 63–69.
Borsi, S. H., Raji, H., Dargahi Malamir, M., Nokhostin, F., Kargaran, A. (2021). Rivaroxaban versus enoxaparin for treatment of patients with nonhematologic cancer with venous thromboembolism a randomized clinial trial. Tehran University Medical Journal TUMS Publications, 79(4), 281–289.
Draghici, S., Khatri, P., Tarca, A. L., Amin, K., Done, A., Voichita, C., et al. (2007). A systems biology approach for pathway level analysis. Genome Research, 17(10), 1537–1545.
Yousefi, S. R., Alshamsi, H. A., Amiri, O., & Salavati-Niasari, M. (2021). Synthesis, characterization and application of Co/Co3O4 nanocomposites as an effective photocatalyst for discoloration of organic dye contaminants in wastewater and antibacterial properties. Journal of Molecular Liquids, 337, 116405.
Mahdi, M. A., Yousefi, S. R., Jasim, L. S., & Salavati-Niasari, M. (2022). Green synthesis of DyBa2Fe3O7.988/DyFeO3 nanocomposites using almond extract with dual eco-friendly applications: Photocatalytic and antibacterial activities. International Journal of Hydrogen Energy, 47(31), 14319–14330.
Hou, Q., Huang, J., Xiong, X., Guo, Y., & Zhang, B. (2022). Role of nutrient-sensing receptor GPRC6A in regulating colonic group 3 innate lymphoid cells and inflamed mucosal healing. Journal of Crohn’s and Colitis, 20, 1–13.
Liu, C., Wang, Y., Li, L., He, D., Chi, J., Li, Q., et al. (2022). Engineered extracellular vesicles and their mimetics for cancer immunotherapy. Journal of Controlled Release, 349, 679–698.
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2012). NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Research, 41(D1), D991–D995.
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research, 43(7), e47.
Carmona-Saez, P., Chagoyen, M., Tirado, F., Carazo, J. M., & Pascual-Montano, A. (2007). GENECODIS: A web-based tool for finding significant concurrent annotations in gene lists. Genome Biology, 8(1), 1–8.
Franz, M., Rodriguez, H., Lopes, C., Zuberi, K., Montojo, J., Bader, G. D., et al. (2018). GeneMANIA update 2018. Nucleic Acids Research, 46(W1), W60–W64.
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P.-L., & Ideker, T. (2011). Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics, 27(3), 431–432.
Chin, C.-H., Chen, S.-H., Wu, H.-H., Ho, C.-W., Ko, M.-T., & Lin, C.-Y. (2014). cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Systems Biology, 8(4), 1–7.
Mizuno, H., Kitada, K., Nakai, K., & Sarai, A. (2009). PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Medical Genomics, 2(1), 1–11.
Firth, J. (2019). Haematology: Multiple myeloma. Clinical Medicine (London), 19(1), 58–60.
Dhodapkar, M. V. (2016). MGUS to myeloma: A mysterious gammopathy of underexplored significance. Blood, 128(23), 2599–2606.
Alipanahzadeh, H., Ghulamreza, R., Shokouhian, M., Bagheri, M., & Maleknia, M. (2020). Deep vein thrombosis: A less noticed complication in hematologic malignancies and immunologic disorders. Journal of Thrombosis and Thrombolysis, 50(2), 318–329.
Rajkumar, S. V. (2020). Multiple myeloma: 2020 Update on diagnosis, risk-stratification and management. American Journal of Hematology, 95(5), 548–567.
Manier, S., Salem, K. Z., Park, J., Landau, D. A., Getz, G., & Ghobrial, I. M. (2017). Genomic complexity of multiple myeloma and its clinical implications. Nature Reviews Clinical Oncology, 14(2), 100–113.
Bianchi, G., & Munshi, N. C. (2015). Pathogenesis beyond the cancer clone(s) in multiple myeloma. Blood, 125(20), 3049–3058.
Rajan, A. M., & Rajkumar, S. V. (2015). Interpretation of cytogenetic results in multiple myeloma for clinical practice. Blood Cancer Journal, 5(10), e365.
Kuehl, W. M., & Bergsagel, P. L. (2002). Multiple myeloma: Evolving genetic events and host interactions. Nature Reviews Cancer, 2(3), 175–187.
Gholami, F., Ghasemi, A., Bahrami, A. R., Bidkhouri, H. R., Mishkin, H. N., Rad, M. P., et al. (2019). The therapeutic effect of autologous bone marrow mesenchymal stem cells to prevent the progress of chronic allograft nephropathy. Journal of Renal Injury Prevention, 8(1), 1–5.
Türkmen, S., Binder, A., Gerlach, A., Niehage, S., Theodora Melissari, M., Inandiklioglu, N., et al. (2014). High prevalence of immunoglobulin light chain gene aberrations as revealed by FISH in multiple myeloma and MGUS. Genes, Chromosomes and Cancer, 53(8), 650–656.
Chretien, M. L., Corre, J., Lauwers-Cances, V., Magrangeas, F., Cleynen, A., Yon, E., et al. (2015). Understanding the role of hyperdiploidy in myeloma prognosis: Which trisomies really matter? Blood, 126(25), 2713–2719.
Saxe, D., Seo, E. J., Bergeron, M. B., & Han, J. Y. (2019). Recent advances in cytogenetic characterization of multiple myeloma. International Journal of Laboratory Hematology, 41(1), 5–14.
Prideaux, S. M., Conway O’Brien, E., & Chevassut, T. J. (2014). The genetic architecture of multiple myeloma. Advances in Hematology, 2014, 864058.
Walker, B. A., Boyle, E. M., Wardell, C. P., Murison, A., Begum, D. B., Dahir, N. M., et al. (2015). Mutational spectrum, copy number changes, and outcome: Results of a sequencing study of patients with newly diagnosed myeloma. Journal of Clinical Oncology, 33(33), 3911–3920.
Moreau, P., & Rajkumar, S. V. (2016). Multiple myeloma—Translation of trial results into reality. Lancet, 388(10040), 111–113.
Fan, F., Bashari, M. H., Morelli, E., Tonon, G., Malvestiti, S., Vallet, S., et al. (2017). The AP-1 transcription factor JunB is essential for multiple myeloma cell proliferation and drug resistance in the bone marrow microenvironment. Leukemia, 31(7), 1570–1581.
Shaulian, E. (2010). AP-1—The Jun proteins: Oncogenes or tumor suppressors in disguise? Cellular Signalling, 22(6), 894–899.
Fan, F., & Podar, K. (2021). The role of AP-1 Transcription factors in plasma cell biology and multiple myeloma pathophysiology. Cancers (Basel), 13(10), 2326.
Atsaves, V., Leventaki, V., Rassidakis, G. Z., & Claret, F. X. (2019). AP-1 transcription factors as regulators of immune responses in cancer. Cancers (Basel), 11(7), 1037.
Cannarile, L., Delfino, D. V., Adorisio, S., Riccardi, C., & Ayroldi, E. (2019). Implicating the role of GILZ in glucocorticoid modulation of T-cell activation. Frontiers in Immunology, 10, 1823.
Grötsch, B., Brachs, S., Lang, C., Luther, J., Derer, A., Schlötzer-Schrehardt, U., et al. (2014). The AP-1 transcription factor Fra1 inhibits follicular B cell differentiation into plasma cells. Journal of Experimental Medicine, 211(11), 2199–2212.
Ubieta, K., Garcia, M., Grötsch, B., Uebe, S., Weber, G. F., Stein, M., et al. (2017). Fra-2 regulates B cell development by enhancing IRF4 and Foxo1 transcription. Journal of Experimental Medicine, 214(7), 2059–2071.
Takayanagi, H., Kim, S., Koga, T., Nishina, H., Isshiki, M., Yoshida, H., et al. (2002). Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Developmental Cell, 3(6), 889–901.
Li, Z., Teng, M., Jiang, Y., Zhang, L., Luo, X., Liao, Y., et al. (2022). YTHDF1 negatively regulates Treponema pallidum-induced inflammation in THP-1 macrophages by promoting SOCS3 translation in an m6A-dependent manner. Frontiers in Immunology, 13, 857727.
Hedvat, C. V., Comenzo, R. L., Teruya-Feldstein, J., Olshen, A. B., Ely, S. A., Osman, K., et al. (2003). Insights into extramedullary tumour cell growth revealed by expression profiling of human plasmacytomas and multiple myeloma. British Journal of Haematology, 122(5), 728–744.
Shen, Y. J., Mishima, Y., Shi, J., Sklavenitis-Pistofidis, R., Redd, R. A., Moschetta, M., et al. (2021). Progression signature underlies clonal evolution and dissemination of multiple myeloma. Blood, 137(17), 2360–2372.
Biran, N., Siegel, D. S., & Vesole, D. H. (2018). The forgotten class of drugs for multiple myeloma: HDAC inhibitors. Lancet Haematology, 5(12), e604–e605.
Kiziltepe, T., Hideshima, T., Catley, L., Raje, N., Yasui, H., Shiraishi, N., et al. (2007). 5-Azacytidine, a DNA methyltransferase inhibitor, induces ATR-mediated DNA double-strand break responses, apoptosis, and synergistic cytotoxicity with doxorubicin and bortezomib against multiple myeloma cells. Molecular Cancer Therapy, 6(6), 1718–1727.
Fan, F., Malvestiti, S., Vallet, S., Lind, J., Garcia-Manteiga, J. M., Morelli, E., et al. (2021). JunB is a key regulator of multiple myeloma bone marrow angiogenesis. Leukemia, 35(12), 3509–3525.
Podar, K., Raab, M. S., Tonon, G., Sattler, M., Barilà, D., Zhang, J., et al. (2007). Up-regulation of c-Jun inhibits proliferation and induces apoptosis via caspase-triggered c-Abl cleavage in human multiple myeloma. Cancer Research, 67(4), 1680–1688.
Chen, L., Wang, S., Zhou, Y., Wu, X., Entin, I., Epstein, J., et al. (2010). Identification of early growth response protein 1 (EGR-1) as a novel target for JUN-induced apoptosis in multiple myeloma. Blood, 115(1), 61–70.
Liu, P., Shi, J., & Wang, Z.-A. (2013). Pattern formation of the attraction–repulsion Keller-Segel system. Discrete and Continuous Dynamical Systems Series B, 18(10), 2597.
Carrasco, D. R., Tonon, G., Huang, Y., Zhang, Y., Sinha, R., Feng, B., et al. (2006). High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell, 9(4), 313–325.
Colla, S., Zhan, F., Xiong, W., Wu, X., Xu, H., Stephens, O., et al. (2007). The oxidative stress response regulates DKK1 expression through the JNK signaling cascade in multiple myeloma plasma cells. Blood, 109(10), 4470–4477.
Fan, F., Tonon, G., Bashari, M. H., Vallet, S., Antonini, E., Goldschmidt, H., et al. (2014). Targeting Mcl-1 for multiple myeloma (MM) therapy: Drug-induced generation of Mcl-1 fragment Mcl-1(128–350) triggers MM cell death via c-Jun upregulation. Cancer Letters, 343(2), 286–294.
Annunziata, C. M., Hernandez, L., Davis, R. E., Zingone, A., Lamy, L., Lam, L. T., et al. (2011). A mechanistic rationale for MEK inhibitor therapy in myeloma based on blockade of MAF oncogene expression. Blood, 117(8), 2396–2404.
Walker, B. A., Mavrommatis, K., Wardell, C. P., Ashby, T. C., Bauer, M., Davies, F. E., et al. (2018). Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood, 132(6), 587–597.
Bergsagel, P. L., & Kuehl, W. M. (2001). Chromosome translocations in multiple myeloma. Oncogene, 20(40), 5611–5622.
Lai, W.-F., Tang, R., & Wong, W.-T. (2020). Ionically crosslinked complex gels loaded with oleic acid-containing vesicles for transdermal drug delivery. Pharmaceutics, 12(8), 725.
Pratt, G., & Morris, T. C. (2017). Review of the NICE guidelines for multiple myeloma. International Journal of Laboratory Hematology, 39(1), 3–13.
Qiang, Y. W., Ye, S., Huang, Y., Chen, Y., Van Rhee, F., Epstein, J., et al. (2018). MAFb protein confers intrinsic resistance to proteasome inhibitors in multiple myeloma. BMC Cancer, 18(1), 724.
Hao, P., Li, H., Zhou, L., Sun, H., Han, J., & Zhang, Z. (2022). Serum metal ion-induced cross-linking of photoelectrochemical peptides and circulating proteins for evaluating cardiac ischemia/reperfusion. ACS Sensors, 7(3), 775–783.
Hideshima, T., Chauhan, D., Richardson, P., Mitsiades, C., Mitsiades, N., Hayashi, T., et al. (2002). NF-kappa B as a therapeutic target in multiple myeloma. Journal of Biological Chemistry, 277(19), 16639–16647.
He, X., Zhu, Y., Yang, L., Wang, Z., Wang, Z., Feng, J., et al. (2021). MgFe-LDH nanoparticles: A promising leukemia inhibitory factor replacement for self-renewal and pluripotency maintenance in cultured mouse embryonic stem cells. Advanced Science, 8(9), 2003535.
Olivier, S., Close, P., Castermans, E., de Leval, L., Tabruyn, S., Chariot, A., et al. (2006). Raloxifene-induced myeloma cell apoptosis: A study of nuclear factor-kappaB inhibition and gene expression signature. Molecular Pharmacology, 69(5), 1615–1623.
Jin, H.-Y., & Wang, Z.-A. (2019). Global stabilization of the full attraction–repulsion Keller–Segel system. arXiv preprint. arXiv:190505990
Yang, W., Liu, W., Li, X., Yan, J., & He, W. (2022). Turning chiral peptides into a racemic supraparticle to induce the self-degradation of MDM2. Journal of Advanced Research. https://doi.org/10.1016/j.jare.2022.05.009
Acknowledgements
The authors appreciate and thank the efforts of the Center for the Development of Clinical Researches of the Educational and Therapeutic Research Complex of Hazrat-e Rasool General Hospital.
Funding
None.
Author information
Authors and Affiliations
Contributions
HR conceived the manuscript and revised it. MM and BSA wrote the manuscript. RM and AH analysis of data. PK conducted the revise. FF conducted the native English.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors. All the procedures performed in the studies involving human participants were in accordance with ethical standards of Local Ethics Committee of Iran University of Medical Sciences (IR.IUMS.REC.1401.607), as well as 1964 Helsinki Declaration.
Informed Consent
This study did not use animal or human samples. Written informed consent was obtained from all patients and normal subjects.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khalili, P., Maddah, R., Maleknia, M. et al. Evaluation of Genes and Molecular Pathways Involved in the Progression of Monoclonal Gammopathy of Undetermined Significance (MGUS) to Multiple Myeloma: A Systems Biology Approach. Mol Biotechnol 65, 1275–1286 (2023). https://doi.org/10.1007/s12033-022-00634-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00634-6