Abstract
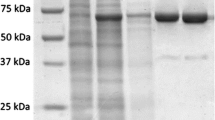
Bacteriophage Phi11 harbors a gene, gp13, encoding the putative SSB protein (GenBank accession no. NC_004615.1). SSB proteins bind to and protect the single-stranded DNA molecules from nuclease digestion and are essential for the growth and metabolic activities of the organisms encoding them. In this investigation, we have carried out the cloning, recombinant expression, and purification of rGp13 for the first time in Escherichia coli. EMSA data indicated that the purified recombinant Gp13 protein was capable of binding to single-stranded DNA. The protein exhibited maximum binding activity at 32 °C. Furthermore, our bioinformatic analysis has revealed that Gp13 consists of an OB-fold, a characteristic of SSB proteins. However, the arrangement of the OB-fold is unique, being located in the C-terminal domain of Gp13. Despite the importance of SSB proteins in various metabolic processes as well as in various types of PCR, there are no reports on the purification and characterization of SSB proteins from staphylococcal bacteriophages. We expect that the purification and characterization of recombinant Gp13 will help us gain a better insight into its biological activity and make it available in large quantities for molecular biology work.








Similar content being viewed by others
Abbreviations
- CD:
-
Conserved Domain
- DNA:
-
Deoxyribonucleic acid
- E. coli:
-
Escherichia coli
- EMSA:
-
Electrophoretic mobility shift assay
- IDT:
-
Integrated DNA Technologies
- IMAC:
-
Immobilized metal affinity chromatography
- IPTG:
-
Isopropyl β-d-1-thiogalactopyranoside
- NCBI:
-
National Center for Biotechnology Information
- Ni–NTA:
-
Nickel Nitrilotriacetic Acid
- OB fold:
-
Oligosaccharide/ oligonucleotide/ oligopeptide binding fold
- PCR:
-
Polymerase Chain Reaction
- PMSF:
-
Phenylmethane sulfonyl fluoride
- RT PCR:
-
Real Time Polymerase Chain Reaction
- S. aureus:
-
Staphylococcus aureus
- SDS-PAGE:
-
Sodium Dodecyl Sulphate–Polyacrylamide Gel Electrophoresis
- SSB:
-
Single-stranded DNA binding protein
References
Wold, M. S. (1997). Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annual Review of Biochemistry, 66, 61–92. https://doi.org/10.1146/annurev.biochem.66.1.61
Lohman, T. M., & Ferrari, M. E. (1994). Escherichia coli single-stranded DNA-binding protein: Multiple DNA-binding modes and cooperativities. Annual Review of Biochemistry, 63, 527–570. https://doi.org/10.1146/annurev.bi.63.070194.002523
Cha, T. A., & Alberts, B. M. (1989). The bacteriophage T4 DNA replication fork: Only DNA helicase is required for leading strand DNA synthesis by the DNA polymerase holoenzyme. Journal of Biological Chemistry, 264(21), 12220–12225.
Kim, Y. T., & Richardson, C. C. (1993). Bacteriophage T7 gene 2.5 protein: An essential protein for DNA replication. Proceedings of the National Academy of Sciences USA, 90(21), 10173–10177. https://doi.org/10.1073/pnas.90.21.10173
Chase, J. W., & Williams, K. R. (1986). Single-stranded DNA binding proteins required for DNA replication. Annual Review of Biochemistry, 55, 103–136. https://doi.org/10.1146/annurev.bi.55.070186.000535
Zou, Y., Liu, Y., Wu, X., & Shell, S. M. (2006). Functions of human replication protein A (RPA): From DNA replication to DNA damage and stress responses. Journal of Cellular Physiology, 208(2), 267–273. https://doi.org/10.1002/jcp.20622
Pestryakov, P. E., & Lavrik, O. I. (2008). Mechanisms of single-stranded DNA-binding protein functioning in cellular DNA metabolism. Biochemistry (Moscow), 73(13), 1388–1404. https://doi.org/10.1134/s0006297908130026
Weiner, J. H., Bertsch, L. L., & Kornberg, A. (1975). The deoxyribonucleic acid unwinding protein of Escherichia coli: Properties and function in replication. Journal of Biological Chemistry, 250(6), 1972–1980. PMID: 1090613.
Olszewski, M., Mickiewicz, M., & Kur, J. (2008). Two highly thermostable paralogous single-stranded DNA-binding proteins from Thermoanaerobacter tengcongensis. Archives of Microbiology, 190(1), 79–87. https://doi.org/10.1007/s00203-008-0366-6
Olszewski, M., Grot, A., Wojciechowski, M., Nowak, M., Mickiewicz, M., & Kur, J. (2010). Characterization of exception ally thermostable single-stranded DNA-binding proteins from Thermotoga maritima and Thermotoga neapolitana. BMC Microbiology, 10, 260. https://doi.org/10.1186/1471-2180-10-260
Bochkarev, A., & Bochkareva, E. (2004). From RPA to BRCA2: Lessons from single-stranded DNA binding by the OB-fold. Current Opinion in Structural Biology, 14(1), 36–42. https://doi.org/10.1016/j.sbi.2004.01.001
Krejci, L., & Sung, P. (2002). RPA not that different from SSB. Structure., 10(5), 601–602. https://doi.org/10.1016/s0969-2126(02)00765-7
Murzin, A. G. (1993). OB (oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO Journal, 12(3), 861–867. PMID: 8458342.
Purnapatre, K., & Varshney, U. (1999). Cloning, over-expression and biochemical characterization of the single-stranded DNA binding protein from mycobacterium tuberculosis. European Journal of Biochemistry, 264(2), 591–598.
Flynn, R. L., & Zou, L. (2010). Oligonucleotide/ oligosaccharide-binding fold proteins: A growing family of genome guardians. Critical Reviews in Biochemistry and Molecular Biology, 45(4), 266–275. https://doi.org/10.3109/10409238.2010.488216
Theobald, D. L., Mitton-Fry, R. M., & Wuttke, D. S. (2003). Nucleic acid recognition by OB-fold proteins. Annual Review of Biophysics and Biomolecular Structure, 32, 115–133. https://doi.org/10.1146/annurev.biophys.32.110601.142506
Perales, C., Cava, F., Meijer, W. J. J., & Berenguer, J. (2003). Enhancement of DNA, cDNA synthesis and fidelity at high temperatures by a dimeric single-stranded DNA-binding protein. Nucleic Acids Research, 31(22), 6473–6480. https://doi.org/10.1093/nar/gkg865
Lohman, T. M., Green, J. M., & Beyer, R. S. (1986). Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product: Expression of the ssb gene under lambda. PL control. Biochemistry, 25(1), 21–25. https://doi.org/10.1021/bi00349a004
Russell, D. W., & Sambrook, J. (2001). Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press.
Lee, C. Y., & Iandolo, J. J. (1986). Integration of staphylococcal phage L54a occurs by site-specific recombination: Structural analysis of the attachment sites. Proceedings of the National academy of Sciences of the United States of America, 83(15), 5474–5478. https://doi.org/10.1073/pnas.83.15.5474
Lee, C., & Iandolo, J. (1988). Structural analysis of staphylococcal bacteriophage phi 11 attachment sites. Journal of Bacteriology, 170(5), 2409–2411. https://doi.org/10.1128/jb.170.5.2409-2411.1988
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., & Struhl, K., Eds. (1998). In current protocols in molecular biology. 2:10.0.1–10.22.24 and 12.0.1–12.2.11 copyright by John Wiley & Sons, Inc.
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., Gwadz, M., Hurwitz, D. I., Lu, F., Marchler, G. H., Song, J. S., Thanki, N., Wang, Z., Yamashita, R. A., Zhang, D., et al. (2017). CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res., 45, 200–203. https://doi.org/10.1093/nar/gku1221
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., Geer, R. C., He, J., Gwadz, M., Hurwitz, D. I., Lanczycki, C. J., Lu, F., Marchler, G. H., Song, J. S., Thanki, N., Wang, Z., Yamashita, R. A., Zhang, D., Zheng, C., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Research, 43, 222–6. https://doi.org/10.1093/nar/gku1221
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., Fong, J. H., Geer, L. Y., Geer, R. C., Gonzales, N. R., Gwadz, M., Hurwitz, D. I., Jackson, J. D., Ke, Z., Lanczycki, C. J., Lu, F., Marchler, G. H., Mullokandov, M., Omelchenko, M. V., et al. (2011). CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Research, 39, 225–9. https://doi.org/10.1093/nar/gkq1189
Marchler-Bauer, A., & Bryant, S. H. (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Research, 32, 327–331. https://doi.org/10.1093/nar/gkh454
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry., 72, 248–254. https://doi.org/10.1006/abio.1976.9999
Das, A., & Biswas, M. (2016). Changes in the functional activity of phi11 cro protein is mediated by various ions. Protein Journal, 35(6), 407–415. https://doi.org/10.1007/s10930-016-9684-8
Hemmadi, V., Das, A., Chouhan, O. P., Biswas, S., & Biswas, M. (2019). Effect of ions and inhibitors on the catalytic activity and structural stability of S aureus enolase. Journal of Biosciences., 44(4), 90. PMID: 31502568.
Micsonai, A., Wien, F., Kernya, L., Lee, Y. H., Goto, Y., Réfrégiers, M., & Kardos, J. (2015). Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proceedings of the National Academy of Sciences USA, 112(24), E3095–E3103.
Micsonai, A., Wien, F., Bulyaki, É., Kun, J., Moussong, É., Lee, Y. H., Goto, Y., Réfrégiers, M., & Kardos, J. (2018). BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Research, 46(W1), W315–W322.
Genschel, J., Curth, U., & Urbanke, C. (2000). Interaction of E. coli single-stranded DNA binding protein (SSB) with exonuclease I: The carboxy-terminus of SSB is the recognition site for the nuclease. Journal of Biological Chemistry, 381(3), 183–192. https://doi.org/10.1515/BC.2000.025
Curth, U., Genschel, J., Urbanke, C., & Greipel, J. (1996). In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Research, 24(14), 2706–2711. https://doi.org/10.1093/nar/24.14.2706
Wadsworth, R. I. M., & White, M. F. (2001). Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Research, 29, 914–920. https://doi.org/10.1093/nar/29.4.914
Meyer, R. R., & Laine, P. S. (1990). The single-stranded DNA-binding protein of Escherichia coli. Microbiological Reviews, 54(4), 342–380. https://doi.org/10.1128/mr.54.4.342-380.1990
Wold, M. S., Mallory, J. B., Roberts, J. D., LeBowitz, J. H., & McMacken, R. (1982). Initiation of bacteriophage lambda DNA replication in vitro with purified lambda replication proteins. Proceedings of the National Academy of Sciences of the United States of America, 79, 6176–6180.
Dodson, M., McMacken, R., & Echols, H. (1989). Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: Protein association and dissociation reactions responsible for localized initiation of replication. Journal of Biological Chemistry, 264, 10719–10725.
Tone, T., Takeuchi, A., & Makino, O. (2012). Single-stranded DNA binding protein Gp5 of Bacillus subtilis phage Φ29 is required for viral DNA replication in growth-temperature dependent fashion. Bioscience, Biotechnology, and Biochemistry, 76(12), 2351–2353. https://doi.org/10.1271/bbb.120587
Hollis, T., Stattel, J. M., Walther, D. S., Richardson, C. C., & Ellenberger, T. (2001). Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophageT7. Proceedings of the National academy of Sciences of the United States of America, 98, 9557–9562152.
Hernandez, A. J., & Richardson, C. C. (2019). Gp2.5, the multifunctional bacteriophageT7 single-stranded DNA binding protein. Seminars in Cell & Developmental Biology, 86, 92–101.
Lee, S. J., Marintcheva, B., Hamdan, S. M., & Richardson, C. C. (2006). The C-terminal residues of bacteriophage T7 gene 4 helicase primase coordinate helicase and DNA polymerase activities. Journal of Biological Chemistry, 281, 25841–25849.
Shereda, R. D., Kozlov, A. G., Lohman, T. M., Cox, M. M., & Keck, J. L. (2008). SSB as an organizer/mobilizer of genome maintenance complexes. Critical Reviews in Biochemistry and Molecular Biology, 43(5), 289–318. https://doi.org/10.1080/10409230802341296
Acknowledgements
I wish to acknowledge the University Grants Commission, New Delhi, for the financial assistance in the form of Junior Research Fellowship (JRF) and Senior Research Fellowship (SRF), which supported the study.
Funding
This work was supported by the financial assistance from the Department of Science and Technology, Govt. of India (Sanction No. CRG/2020/004968).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
There is no conflict of interest regarding the publication of this paper.
Ethical Approval
This article does not contain descriptions of studies performed by the authors with the participation of humans or using animals as objects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ratre, V., Hemmadi, V., Biswas, S. et al. Identification and Preliminary Characterization of a Novel Single-Stranded DNA Binding Protein of Staphylococcus aureus Phage Phi11 Expressed in Escherichia coli. Mol Biotechnol 65, 922–933 (2023). https://doi.org/10.1007/s12033-022-00598-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00598-7