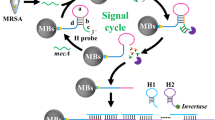
Abstract
Despite major advances in combating pathogenic bacteria caused infection, infectious diseases remain a major cause of death today. A variety of bacteria detection approaches have been established by exploiting the aptamer to specifically identify target. However, these methods require specific equipment or laboratory based instruments, which is expensive and not easily available to the public. To deal with this issue, we develop here a portable and sensitive pathogenic bacteria detection approach through integrating capture probe based recognition and Exo-III assisted signal amplification. In this method, a designed capture probe identifies the surface protein of target bacteria, leading to the release of blocker sequence. The released blocker sequence mediates Exo-III enzyme based signal amplification, generating a large amount of signal strand DNA (ssDNA) sequences. The obtained ssDNA works as a linker to conjugate streptavidin magnetic beads (SMBs) with DNA invertase. After magnet based enrichment and washing away of free DNA invertase, the SMBs-DNA invertase complex is used to catalyze the hydrolysis of sucrose into glucose for PGM readout. Based on this, the approach exhibits a 6 order of magnitudes in detecting Staphylococcus Aureus (S. aureus) with the limit of detection as low as 78 cfu/mL. In addition, the capability to specifically detect target bacteria and high repeatability (relative standard deviation, 4.44%) endows the method wider applicable scenes. In all, the established shows a promising prospect in the detection of bacteria pathogen and the early-diagnosis of bacterial infections.
Graphical Abstract





Similar content being viewed by others
Data Availability
Almost all details of experimental data are presented in the article or additional file.
References
Lancellotti, M., Pereira, R. F., Cury, G. G., & Hollanda, L. M. (2009). Pathogenic and opportunistic respiratory bacteria-induced apoptosis. The Brazilian Journal of Infectious Diseases, 13(3), 226–231.
Xue, T., Lu, Y., Yang, H., Hu, X., Zhang, K., Ren, Y., Wu, C., Xia, X., Deng, R., & Wang, Y. (2022). Isothermal RNA amplification for the detection of viable pathogenic bacteria to estimate the salmonella virulence for causing enteritis. Journal of Agriculture and Food Chemistry, 70(5), 1670–1678.
Langford, B. J., So, M., Raybardhan, S., Leung, V., Westwood, D., MacFadden, D. R., Soucy, J. R., & Daneman, N. (2020). Bacterial co-infection and secondary infection in patients with COVID-19: A living rapid review and meta-analysis. Clinical Microbiology & Infection, 26(12), 1622–1629.
Martin, R. M., & Bachman, M. A. (2018). Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Frontiers in Cellular and Infection Microbiology, 8, 4.
Chen, J., Xu, Y., Yan, H., Zhu, Y., Wang, L., Zhang, Y., Lu, Y., & Xing, W. (2018). Sensitive and rapid detection of pathogenic bacteria from urine samples using multiplex recombinase polymerase amplification. Lab on a Chip, 18(16), 2441–2452.
Pala, L., Sirec, T., & Spitz, U. (2020). Modified enzyme substrates for the detection of bacteria: A review. Molecules, 25(16), 3960.
Rajapaksha, P., Elbourne, A., Gangadoo, S., Brown, R., Cozzolino, D., & Chapman, J. (2019). A review of methods for the detection of pathogenic microorganisms. The Analyst, 144(2), 396–411.
Lee, K. J., Lee, W. S., Hwang, A., Moon, J., Kang, T., Park, K., & Jeong, J. (2017). Simple and rapid detection of bacteria using a nuclease-responsive DNA probe. The Analyst, 143(1), 332–338.
Maldonado, J., Estevez, M. C., Fernandez-Gavela, A., Gonzalez-Lopez, J. J., Gonzalez-Guerrero, A. B., & Lechuga, L. M. (2020). Label-free detection of nosocomial bacteria using a nanophotonic interferometric biosensor. The Analyst, 145(2), 497–506.
Norouz, D. A., Simsek Ozek, N., Aysin, F., Calis, A., Yilmaz, A., & Yilmaz, M. (2021). Combining vancomycin-modified gold nanorod arrays and colloidal nanoparticles as a sandwich model for the discrimination of gram-positive bacteria and their detection via surface-enhanced Raman spectroscopy (SERS). The Analyst, 146(11), 3642–3653.
Qiao, T., Kim, S., Lee, W., & Lee, H. (2021). An enhanced fluorescence detection of a nitroaromatic compound using bacteria embedded in porous poly lactic-co-glycolic acid microbeads. The Analyst, 146(14), 4615–4621.
Xu, L., Dai, Q., Shi, Z., Liu, X., Gao, L., Wang, Z., Zhu, X., & Li, Z. (2020). Accurate MRSA identification through dual-functional aptamer and CRISPR-Cas12a assisted rolling circle amplification. Journal of Microbiol Methods, 173, 105917.
Ahn, J. K., Kim, H. Y., Park, K. S., & Park, H. G. (2018). A personal glucose meter for label-free and washing-free biomolecular detection. Analytical Chemistry, 90(19), 11340–11343.
Li, F., Li, X., Zhu, N., Li, R., Kang, H., & Zhang, Q. (2020). An aptasensor for the detection of ampicillin in milk using a personal glucose meter. Analytical Methods, 12(26), 3376–3381.
Zhang, H., Chen, G. Y., Qian, Z. M., Li, W. J., Li, C. H., Hu, Y. J., & Yang, F. Q. (2021). A portable personal glucose meter method for enzyme activity detection and inhibitory activity evaluation based on alkaline phosphatase-mediated reaction. Analytical and Bioanalytical Chemistry, 413(9), 2457–2466.
Xue-tao, X., Kai-yi, L., & Jia-ying, Z. (2014). Portable and sensitive quantitative detection of DNA using personal glucose meters and exonuclease III-assisted signal amplification. The Analyst, 139(19), 4982–4986.
Kim, D. M., & Yoo, S. M. (2020). DNA-modifying enzyme reaction-based biosensors for disease diagnostics: Recent biotechnological advances and future perspectives. Critical Reviews in Biotechnology, 40(6), 787–803.
Liu, H., You, Y., Zhu, Y., & Zheng, H. (2021). Recent advances in the exonuclease III-assisted target signal amplification strategy for nucleic acid detection. Analytical Methods, 13(43), 5103–5119.
Zhao, X., Yuan, Y., Liu, X., Mao, F., Xu, G., & Liu, Q. (2022). A versatile platform for sensitive and label-free identification of biomarkers through an Exo-III-assisted cascade signal amplification strategy. Analytical Chemistry, 94(4), 2298–2304.
Li, Y., Xu, F., Zhang, J., Huang, J., Shen, D., Ma, Y., Wang, X., Bian, Y., & Chen, Q. (2021). Sensitive and label-free detection of bacteria in osteomyelitis through Exo III-assisted cascade signal amplification. ACS Omega, 6(18), 12223–12228.
Dong, N., Jiang, N., Zhao, J., Zhao, G., & Wang, T. (2022). Sensitive and enzyme-free pathogenic bacteria detection through self-circulation of molecular beacon. Applied Biochemistry and Biotechnology, 194(8), 3668–3676.
Acknowledgements
The authors thank financial and technical support from Ningbo Yinzhou No. 2 Hospital.
Funding
No fund available.
Author information
Authors and Affiliations
Contributions
QW is the supervisor of the team in all research steps including designing, data analysis and manuscript writing. XW, as the first author, has the main role for experimental data collection, data gathering, preparation of results, and data analysis. JL, JZ, and YW assist the data analysis.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Ethical Approval
The manuscript does not contain clinical or trial studies on patients, humans, or animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Weng, X., Lou, J., Zhang, J. et al. Sensitive and Portable Detection of Bacteria Using Exonuclease-III (Exo-III) Assisted Signal Amplification and Personal Glucose Meters. Mol Biotechnol 65, 934–941 (2023). https://doi.org/10.1007/s12033-022-00597-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00597-8