Abstract
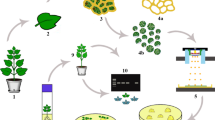
Epigenetic regulation by histone modification can activate or repress transcription through changes in chromatin dynamics and regulates development and the response to environmental signals in both animals and plants. Chromatin immunoprecipitation (ChIP) is an indispensable tool to identify histones with specific post-translational modifications. The lack of a ChIP technique for macroalgae has hindered understanding of the role of histone modification in the expression of genes in this organism. In this study, a ChIP method with several modifications, based on existing protocols for plant cells, has been developed for the red macroalga, Neopyropia yezoensis, that consists of a heterogeneous alternation of macroscopic leaf-like gametophytes and microscopic filamentous sporophytes. ChIP method coupled with qPCR enables the identification of a histone mark in generation-specific genes from N. yezoensis. The results indicate that acetylation of histone H3 at lysine 9 in the 5′ flanking and coding regions from generation-specific genes was maintained at relatively high levels, even in generation-repressed gene expression. The use of this ChIP method will contribute significantly to identify the epigenetic regulatory mechanisms through histone modifications that control a variety of biological processes in red macroalgae.






Similar content being viewed by others
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Jenuwein, T., & Allis, C. D. (2001). Translating the histone code. Science, 293, 1074–1080.
Allis, C. D., & Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. Nature Reviews Genetics, 17, 487–500.
Onufriev, A. V., & Schiessel, H. (2019). The nucleosome: From structure to function through physics. Current Opinion in Structural Biology, 56, 119–130.
Teperino, R., Schoonjans, K., & Auwerx, J. (2010). Histone methyl transferases and demethylases; Can they link metabolism and transcription? Cell Metabolism, 12, 321–327.
Bourdareau, S., Tirichine, L., Lombard, B., Loew, D., Scornet, D., Wu, Y., Coelho, S. M., & Cock, J. M. (2021). Histone modifications during the life cycle of the brown alga Ectocarpus. Genome Biology, 22, 12.
Nelson, J. D., Denisenko, O., & Bomsztyk, K. (2006). Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nature Protocols, 1, 179–185.
Orlando, V. (2000). Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends in Biochemical Sciences, 25, 99–104.
Huang, X. R., Pan, Q. W., Lin, Y., Gu, T. T., & Li, Y. (2020). A native chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in strawberry fruits. Plant Methods. https://doi.org/10.1186/s13007-020-0556-z
O’neill, L. P., & Turner, B. M. (2003). Immunoprecipitation of native chromatin: NChIP. Methods, 31, 76–82.
Drew, K. (1949). Conchocelis-phase in the life history of Porphyra umbilicalis (L.) Kütz. Nature, 164, 748–749.
Iwasaki, H. (1961). The life cycle of Porphyra tenera in vitro. Biological Bulletin of the Marine Biological Laboratory, Woods Hole, 121, 173–187.
Kurogi, M. (1953). Studies of the life history of Porphyra. I. The germination and development of carpospores. Bulletin of Tohoku Regional Fisheries Research Laboratory, 2, 67–103.
Luo, Q., Zhu, Z., Zhu, Z., Yang, R., Qian, F., Chen, H., & Yan, X. (2014). Different responses to heat shock stress revealed heteromorphic adaptation strategy of Pyropia haitanensis (Bangiales, Rhodophyta). PLoS ONE. https://doi.org/10.1371/journal.pone.0094354
Uji, T., Gondaira, Y., Fukuda, S., Mizuta, H., & Saga, N. (2019). Characterization and expression profiles of small heat shock proteins in the marine red alga Pyropia yezoensis. Cell Stress and Chaperones, 24, 223–233.
Inoue, A., Mashino, C., Uji, T., Saga, N., Mikami, K., & Ojima, T. (2015). Characterization of an eukaryotic PL-7 alginate lyase in the marine red alga Pyropia yezoensis. Current Biotechnology, 4, 240–248.
Matsuda, R., Ozgur, R., Higashi, Y., Takechi, K., Takano, H., & Takio, S. (2015). Preferential expression of a bromoperoxidase in sporophytes of a red alga, Pyropia yezoensis. Marine Biotechnology, 17, 199–210.
Uji, T., Hirata, R., Mikami, K., Mizuta, H., & Saga, N. (2012). Molecular characterization and expression analysis of sodium pump genes in the marine red alga Porphyra yezoensis. Molecular Biology Reports, 39, 7973–7980.
Uji, T., Mizuta, H., & Saga, N. (2013). Characterization of the sporophyte-preferential gene promoter from the red alga Porphyra yezoensis using transient gene expression. Marine Biotechnology, 15, 188–196.
Uji, T., Ueda, S., & Mizuta, H. (2022). Identification, characterization, and expression analysis of spondin-like and fasciclin-like genes in Neopyropia yezoensis, a marine red alga. Phycology, 2, 45–59.
Chan, C. X., Blouin, N. A., Zhuang, Y., Zäuner, S., Prochnik, S. E., Lindquist, E., Lin, S., Benning, C., Lohr, M., Yarish, C., Gantt, E., Grossman, A. R., Lu, S., Müller, K. W., Stiller, J., Brawley, S. H., & Bhattacharya, D. (2012). Porphyra (Bangiophyceae) transcriptomes provide insights into red algal development and metabolism. Journal of Phycology, 48, 1328–1342.
Butterfield, N. J. (2000). Bangiomorpha pubescens n. gen., n. sp.: Implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology, 26, 386–404.
Sutherland, J. E., Lindstrom, S. C., Nelson, W. A., Brodie, J., Lynch, M. D. J., Hwang, M. S., Choi, H.-G., Miyata, M., Kikuchi, N., Oliveira, M. C., Farr, T., Neefus, C., Mols-Mortensen, A., Milstein, D., & Müller, K. M. (2011). A new look at an ancient order: Generic revision of the Bangiales (Rhodophyta). Journal of Phycology, 47, 1131–1151.
Saleh, A., Alvarez-Venegas, R., & Avramova, Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nature Protocols, 3, 1018–1025.
Kuwano, K., Aruga, Y., & Saga, N. (1996). Cryopreservation of clonal gametophytic thalli of Porphyra (Rhodophyta). Plant Science., 116, 117–124.
Provasoli, L. (1996). Media and prospects for the cultivation of marine algae. In: A. Watanabe, & A. Hattori (Eds.), Culture and collections of algae, Proc U S-Japan Conf, Hakone, Jpn Soc Plant Physiol, Tokyo, 1968 (pp. 63–75)
Uji, T., Matsuda, R., Takechi, K., Takano, H., Mizuta, H., & Takio, S. (2016). Ethylene regulation of sexual reproduction in the marine red alga Pyropia yezoensis (Rhodophyta). Journal of Applied Phycology, 28, 3501–3509.
Ali, S., Khan, N., & Tang, Y. (2022). Epigenetic marks for mitigating abiotic stresses in plants. Journal of Plant Physiology, 275, 153740.
Loidl, P. (2004). A plant dialect of the histone language. Trends in Plant Science, 9, 84–90. https://doi.org/10.1016/j.tplants.2003.12.007
Vastenhouw, N. L., & Schier, A. F. (2012). Bivalent histone modifications in early embryogenesis. Current Opinion in Cell Biology, 24, 374–386.
Kurita, K., Sakamoto, T., Yagi, N., Sakamoto, Y., Ito, A., Nishino, N., Sako, K., Yoshida, M., Kimura, H., Seki, M., & Matsunaga, S. (2017). Live imaging of H3K9 acetylation in plant cells. Scientific Reports, 7, 45894.
Zhou, J., Wang, X., He, K., Charron, J. B., Elling, A. A., & Deng, X. W. (2010). Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Molecular Biology, 72, 585–595.
Wei, L., & Xu, J. (2018). Optimized methods of chromatin immunoprecipitation for profiling histone modifications in industrial microalgae Nannochloropsis spp. Journal of Phycology., 54, 358–367.
Kominami, S., Mizuta, H., & Uji, T. (2022). Transcriptome profiling in the marine red alga Neopyropia yezoensis under light/dark cycle. Marine Biotechnology, 24, 393–407.
Hennig, L., Bouveret, R., & Gruissem, W. (2005). MSI1-like proteins: An escort service for chromatin assembly and remodeling complexes. Trends in Cell Biology, 15, 295–302.
Rea, S., Eisenhaber, F., O’Carroll, D., Strahl, B. D., Sun, Z. W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C. P., Allis, C. D., & Jenuwein, T. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406, 593–599.
Ahmad, A., Zhang, Y., & Cao, X. F. (2010). Decoding the epigenetic language of plant development. Molecular Plant, 3, 719–728.
Skvortsova, K., Iovino, N., & Bogdanović, O. (2018). Functions and mechanisms of epigenetic inheritance in animals. Nature Reviews Molecular Cell Biology, 19, 774–790.
Yu, C., Xu, K., Wang, W., Xu, Y., Ji, D., Chen, C., & Xie, C. (2018). Detection of changes in DNA methylation patterns in Pyropia haitanensis under high-temperature stress using a methylation-sensitive amplified polymorphism assay. Journal of Applied Phycology, 30, 2091–2100.
Cao, M., Wang, D., Kong, F., Wang, J., Xu, K., & Mao, Y. (2019). A genome-wide identification of osmotic stress-responsive microRNAs in Pyropia haitanensis (Bangiales, Rhodophyta). Frontiers in Marine Science, 6, 766.
He, L., Huang, A., Shen, S., Niu, J., & Wang, G. (2012). Comparative analysis of microRNAs between sporophyte and gametophyte of Porphyra yezoensis. Comparative and Functional Genomics. https://doi.org/10.1155/2012/912843
Acknowledgements
We wish to thank Drs. Katsutoshi Arai, Takafumi Fujimoto, and Toshiya Nishimura (Hokkaido University, Japan) for kindly providing the LightCycler 480 system.
Funding
This work was supported by the Grant-in-Aid for Young Scientists [Grant Number 19K15907 and 22K05779 to TU] from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ueda, S., Mizuta, H. & Uji, T. Development of Chromatin Immunoprecipitation for the Analysis of Histone Modifications in Red Macroalga Neopyropia yezoensis (Rhodophyta). Mol Biotechnol 65, 590–597 (2023). https://doi.org/10.1007/s12033-022-00562-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00562-5