Abstract
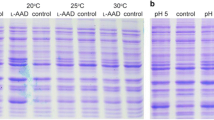
This study reported the cloning, expression, and characterization of a new salt-tolerant leucine dehydrogenase (PrLeuDH) from Pseudoalteromonas rubra DSM 6842. A codon-optimized 1038 bp gene encoding PrLeuDH was successfully expressed on pET-22b( +) in E. coli BL21(DE3). The purified recombinant PrLeuDH showed a single band of about 38.7 kDa on SDS-PAGE. It exhibited the maximum activity at 40 °C and pH 10.5, while kept high activities in the range of 25–45 °C and pH 9.5–12. The Km value and turnover number kcat for leucine of PrLeuDH were 2.23 ± 0.12 mM and 35.39 ± 0.05 s−1, respectively, resulting in a catalytic efficiency kcat/Km of 15.87 s−1/mM. Importantly, PrLeuDH remained 92.1 ± 2.67% active in the presence of 4.0 M NaCl. The study provides the first in-depth understanding of LeuDH from marine Pseudoalteromonas rubra, meanwhile the unique properties of high activity at low temperature and high salt tolerance make it a promising biocatalyst for the synthesis of non-protein amino acids and α-ketoacids under special conditions in pharmaceutical industry.








Similar content being viewed by others
References
Polaina, J., & Maccabe, A. P. (2007). Industrial enzymes. Amino Acid Dehydrogenases., 10, 489–504.
Brunhuber, N., & Blanchard, J. S. (1994). The biochemistry and enzymology of amino acid dehydrogenases. Critical Reviews in Biochemistry & Molecular Biology., 29, 415–467.
Inoue, K., Makino, Y., & Itoh, N. (2005). Production of (R)-chiral alcohols by a hydrogen-transfer bioreduction with NADH-dependent Leifsonia alcohol dehydrogenase (LSADH). ChemInform, 16, 2539–2549.
Sanwal, B. D., & Zink, M. W. (1961). L-Leucine dehydrogenase of Bacillus cereus. Archives of Biochemistry & Biophysics., 94, 430–435.
Zink, M. W., & Sanwal, B. D. (1962). The distribution and substrate specificity of -leucine dehydrogenase. Archives of Biochemistry & Biophysics., 99, 72–77.
Zhu, W., Li, Y., Jia, H., Wei, P., Zhou, H., & Jiang, M. (2016). Expression, purification and characterization of a thermostable leucine dehydrogenase from the halophilic thermophile Laceyella sacchari. Biotechnology Letters., 38, 855–861.
Ohshima, T., Nishida, N., Bakthavatsalam, S., Kataoka, K., Takada, H., Yoshimura, T., Esaki, N., & Soda, K. (2010). The purification, characterization, cloning and sequencing of the gene for a halostable and thermostable leucine dehydrogenase from Thermoactinomyces intermedius. Febs Journal., 222, 305–312.
Nagata, S., Bakthavatsalam, S., & Galkin…, A. (1995). Gene cloning, purification, and characterization of thermostable and halophilic leucine dehydrogenase from a halophilic thermophile, Bacillus licheniformis TSN9. Applied Microbiology and Biotechnology., 44, 432–438.
Reina, K., Shinji, N., Akira, O., & Ohshima, T. (2003). Purification and characterization of leucine dehydrogenase from an alkaliphilic halophile, Natronobacterium magadii MS-3. Journal of Molecular Catalysis B Enzymatic., 23, 231–238.
Xavier, R., & Patrice, G. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research, 42, W320.
Watts, A., Sankaranarayanan, S., Watts, A., & Raipuria, R. K. (2021). Optimizing protein expression in heterologous system: Strategies and tools. Meta Gene., 29, 100899.
Xie, B. B., Shu, Y. L., Qin, Q. L., Rong, J. C., Zhang, X. Y., Chen, X. L., Zhou, B. C., & Zhang, Y. Z. (2012). Genome sequence of the cycloprodigiosin-producing bacterial strain Pseudoalteromonas rubra ATCC 29570T. Journal of Bacteriology, 194, 1637–1638.
Baker, P. J., Turnbull, A. P., Sedelnikova, S. E., Stillman, T. J., & Rice, D. W. (1995). A role for quaternary structure in the substrate specificity of leucine dehydrogenase. Structure, 3, 693–705.
Andrew, W., Martino, B., Stefan, B., Gabriel, S., Gerardo, T., Rafal, G., Heer, F. T., de Beer, A. P., Christine, R., & Lorenza, B. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Research, 46, W296–W303.
Gabriel, S., Christine, R., Waterhouse, A. M., Rafal, G., Juergen, H., & Torsten, S. (2019). QMEAND is co–distance constraints applied on model quality estimation. Bioinformatics, 36, 1765–1771.
Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., & Richardson, D. C. (2010). MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D Biological Crystallography., 66, 12–21.
Nakasako, M., Fujisawa, T., Adachi, S., Kudo, T., & Higuchi, S. (2001). Large-scale domain movements and hydration structure changes in the active-site cleft of unligated glutamate dehydrogenase from Thermococcus profundus studied by cryogenic X-ray crystal structure analysis and small-angle X-ray scattering. Biochemistry, 40, 3069.
Oliveira, T., Sharkey, M. A., Engel, P. C., & Khan, A. R. (2016). Crystal structure of a chimaeric bacterial glutamate dehydrogenase. Acta Crystallographica Section F: Structural Biology Communications., 72, 462–466.
Grzechowiak, M., Sliwiak, J., Ja Skolski, M., & Ruszkowski, M. (2020). Structural studies of glutamate dehydrogenase (Isoform 1) From Arabidopsis thaliana, an important enzyme at the branch-point between carbon and nitrogen metabolism. Frontiers in Plant Science., 11, 1.
Yamaguchi, H., Kamegawa, A., Nakata, K., Kashiwagi, T., Mizukoshi, T., Fujiyoshi, Y., & Tani, K. (2018). Structural insights into thermostabilization of leucine dehydrogenase from its atomic structure by cryo-electron microscopy. Journal of Structural Biology, 205, 11–21.
Zhao, Y., Wakamatsu, T., Doi, K., Sakuraba, H., & Ohshima, T. (2013). Studies on enzymatic properties and crystal structure of L-leucine dehydrogenase from a psychrophilic bacterium Sporosarcina psychrophila. Journal of Molecular Catalysis B Enzymatic, 83, 65–72.
Zhao, Y., Wakamatsu, T., Doi, K., Sakuraba, H., & Ohshima, T. (2012). A psychrophilic leucine dehydrogenase from Sporosarcina psychrophila: Purification, characterization, gene sequencing and crystal structure analysis. Journal of Molecular Catalysis B: Enzymatic., 83, 65–72.
Yamaguchi, H., Kamegawa, A., Nakata, K., Kashiwagi, T., Mizukoshi, T., Fujiyoshi, Y., & Tani, K. (2018). Structural insights into thermostabilization of leucine dehydrogenase from its atomic structure by cryo-electron microscopy. Journal of Structural Biology., 205, 11–21.
Mahdizadehdehosta, R., Kianmehr, A., & Khalili, A. (2013). Isolation and characterization of Leucine dehydrogenase from a thermophilic Citrobacter freundii JK-91strain Isolated from Jask Port. Iranian Journal of Microbiology., 5, 278–284.
Jiang, W., Sun, D., Lu, J., Wang, Y., Wang, S., Zhang, Y., & Fang, B. (2016). A cold-adapted leucine dehydrogenase from marine bacterium Alcanivorax dieselolei: Characterization and l-tert-leucine production. Engineering in Life Sciences., 16, 283–289.
Wang, Y., Hou, Y., Yifan Wang, L., Zheng, X. X., Pan, K., Li, R., & Wang, Q. (2018). A novel cold-adapted leucine dehydrogenase from antarctic sea-ice bacterium Pseudoalteromonas sp. ANT178. Marine Drugs., 16, 359.
Lu, J., Zhang, Y., Sun, D., Jiang, W., Wang, S., & Fang, B. (2016). the development of leucine dehydrogenase and formate dehydrogenase bifunctional enzyme cascade improves the biosynthsis of L-tert-leucine. Applied Biochemistry & Biotechnology., 180, 1–16.
Karst, U., Schutte, H., Baydoun, H., & Tsai, H. (1989). Purification and characterization of leucine dehydrogenase from the thermophile “Bacillus caldolyticus.” Microbiology, 16, 210–216.
Oren, A. (2008). Microbial life at high salt concentrations: Phylogenetic and metabolic diversity. Saline Systems., 4, 1–13.
Graziano, G., & Merlino, A. (2014). Molecular bases of protein halotolerance. Biochimica et Biophysica Acta—Proteins & Proteomics, 1844, 850–858.
Kastritis, P. L., Papandreou, N. C., & Hamodrakas, S. J. (2007). Haloadaptation: Insights from comparative modeling studies of halophilic archaeal DHFRs. International Journal of Biological Macromolecules., 41, 447–453.
Pieper, U., Kapadia, G., Mevarech, M., & Herzberg, O. (1998). Structural features of halophilicity derived from the crystal structure of dihydrofolate reductase from the Dead Sea halophilic archaeon, Haloferax volcanii. Structure., 6, 75–88.
Tadeo, X., López-Méndez, B., Trigueros, T., Laín, A., Casta, O. D., Millet, O., & Petsko, G. A. (2009). Structural basis for the aminoacid composition of proteins from halophilic archea. Plos Biology., 7, e1000257.
Li, F. L., Shi, Y., Zhang, J. X., Gao, J., & Zhang, Y. W. (2018). Cloning, expression, characterization and homology modeling of a novel water-forming NADH oxidase from Streptococcus mutans ATCC 25175. International Journal of Biological Macromolecules, 113, 1073–1079.
Galkin, A., Kulakova, L., Ashida, H., Sawa, Y., & Esaki, N. (1999). Cold-adapted alanine dehydrogenases from two Antarctic bacterial strains: Gene cloning, protein characterization, and comparison with mesophilic and thermophilic counterparts. Applied and Environmental Microbiology., 65, 4014–4020.
Siglioccolo, A., Gerace, R., & Pascarella, S. (2010). “Cold spots” in protein cold adaptation: Insights from normalized atomic displacement parameters (B′ -factors). Biophysical Chemistry., 153, 104–114.
Acknowledgements
This work was supported by the Guangxi science and technology base and talent special project (No. AD211064) and the Beihai "13th five year plan" marine economic innovation and development demonstration project (No. Bhsfs010-4).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest declared.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, R., Liao, YT., Gao, TT. et al. Novel Salt-Tolerant Leucine Dehydrogenase from Marine Pseudoalteromonas rubra DSM 6842. Mol Biotechnol 64, 1270–1278 (2022). https://doi.org/10.1007/s12033-022-00505-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00505-0