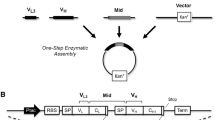
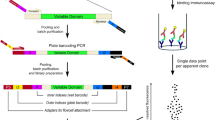
Abstract
Current developments in meta-data analysis and predictive computational models offer alternative routes for the identification of antibodies. In silico-based technologies and NGS data analysis from Ab phage-display selections offer expanded selections of Ab candidates. Accordingly, the identified de novo Abs with predicted selectivity for a target antigen must undergo rapid gene synthesis for downstream Ab characterizations. Here we describe a high-throughput strategy for the generation of synthetic Ab clones for expression as Fab proteins in Escherichia coli. Our approach utilizes simultaneous single-stranded site-directed mutagenesis of diversified Ab regions of a phagemid template with engineered complementary determining regions that contain multiple stop codon and restriction enzyme sites. Subsequently, we perform rapid screening of Ab DNA clones for correct gene assemblies by high-throughput Ab-phage protein expression screens. Identified sequences are corroborated by Sanger DNA sequencing analysis. In summary, our work describes a rapid and cost-effective platform for the high-throughput synthesis of synthetic Ab genes as Fab proteins for implementation into downstream protein validation pipelines.



Similar content being viewed by others
Abbreviations
- Abs:
-
Antibodies
- CDRs:
-
Complementary determining regions
- NGS:
-
Next-generation sequencing
- IPTG:
-
Isopropyl β-d-1-thiogalactopyranoside
- PBS:
-
Phosphate-buffered saline
- ssDNA:
-
Single-stranded DNA
- TMAC:
-
Tetramethyl-ammonium-chloride
- DTT:
-
Dithiothreitol
- BSA:
-
Bovine serum albumin
- HRP:
-
Horseradish peroxidase
References
Feldhaus, M. J., & Siegel, R. W. (2004). Yeast display of antibody fragments: A discovery and characterization platform. Journal of Immunological Methods, 290(1–2), 69–80. https://doi.org/10.1016/J.JIM.2004.04.009.
Hanes, J., Schaffitzel, C., Knappik, A., & Plückthun, A. (2000). Picomolar affinity antibodies from a fully synthetic naive library selected and evolved by ribosome display. Nature Biotechnology, 18(12), 1287–1292. https://doi.org/10.1038/82407.
Winter, G., & Milstein, C. (1991). Man-made antibodies. Nature, 349(6307), 293–299. https://doi.org/10.1038/349293a0.
Marks, J. D., Hoogenboom, H. R., Bonnert, T. P., McCafferty, J., Griffiths, A. D., & Winter, G. (1991). By-passing immunization. Journal of Molecular Biology, 222(3), 581–597. https://doi.org/10.1016/0022-2836(91)90498-U.
Goding, J. W. (1980). Antibody production by hybridomas. Journal of Immunological Methods, 39(4), 285–308. https://doi.org/10.1016/0022-1759(80)90230-6.
Tang, D. C., DeVit, M., & Johnston, S. A. (1992). Genetic immunization is a simple method for eliciting an immune response. Nature, 356(6365), 152–154. https://doi.org/10.1038/356152a0.
Köhler, G., & Milstein, C. (1975). Continuous cultures of fused cells secreting antibody of predefined specificity. Nature, 256, 495–497. https://doi.org/10.1038/256495a0.
Sidhu, S. S. (2005). Phage display in biotechnology and drug discovery. Boca Raton: CRC Press. https://doi.org/10.1201/9780849359125.
Bradbury, A. R. M., & Marks, J. D. (2004). Antibodies from phage antibody libraries. Journal of Immunological Methods, 290(1–2), 29–49. https://doi.org/10.1016/j.jim.2004.04.007.
Yang, G.-H., Yoon, S. O., Jang, M. H., & Hong, H. J. (2007). Affinity maturation of an anti-hepatitis B virus PreS1 humanized antibody by phage display. Journal of microbiology, 45(6), 528–533.
Bradbury, A. R. M., Sidhu, S., Dübel, S., & McCafferty, J. (2011). Beyond natural antibodies: the power of in vitro display technologies. Nature Biotechnology, 29(3), 245–254. https://doi.org/10.1038/nbt.1791.
Miersch, S., & Sidhu, S. S. (2012). Synthetic antibodies: Concepts, potential and practical considerations. Methods, 57(4), 486–498. https://doi.org/10.1016/j.ymeth.2012.06.012.
Benhar, I. (2007). Design of synthetic antibody libraries. Expert Opinion on Biological Therapy, 7(5), 763–779. https://doi.org/10.1517/14712598.7.5.763.
Birtalan, S., Zhang, Y., Fellouse, F. A., Shao, L., Schaefer, G., & Sidhu, S. S. (2008). The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. Journal of Molecular Biology, 377(5), 1518–1528. https://doi.org/10.1016/j.jmb.2008.01.093.
Glanville, J., Zhai, W., Berka, J., Telman, D., Huerta, G., Mehta, G. R., et al. (2009). Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proceedings of the National Academy of Sciences, 106(48), 20216–20221. https://doi.org/10.1073/pnas.0909775106.
Das, R., Baker, D., Nussinov, R., Wolfson, H. J., Corn, J. E., Strauch, E.-M., et al. (2008). Macromolecular modeling with rosetta. Annual Review of Biochemistry, 77(4), 363–382. https://doi.org/10.1146/annurev.biochem.77.062906.171838.
Fleishman, S. J., Whitehead, T. A., Ekiert, D. C., Dreyfus, C., Corn, J. E., Strauch, E.-M., et al. (2011). Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science, 332(6031), 816–821. https://doi.org/10.1126/science.1202617.
Huang, P.-S., Love, J. J., & Mayo, S. L. (2007). A de novo designed protein–protein interface. Protein Science, 16(12), 2770–2774. https://doi.org/10.1110/ps.073125207.
Kries, H., Blomberg, R., & Hilvert, D. (2013). De novo enzymes by computational design. Current Opinion in Chemical Biology, 17(2), 221–228. https://doi.org/10.1016/j.cbpa.2013.02.012.
Der, B. S., & Kuhlman, B. (2011). From computational design to a protein that binds. Science, 332(6031), 801–802. https://doi.org/10.1126/science.1207082.
Kortemme, T., Joachimiak, L. A., Bullock, A. N., Schuler, A. D., Stoddard, B. L., & Baker, D. (2004). Computational redesign of protein-protein interaction specificity. Nature Structural & Molecular Biology, 11(4), 371–379. https://doi.org/10.1038/nsmb749.
Lazar, G. A., Dang, W., Karki, S., Vafa, O., Peng, J. S., Hyun, L., et al. (2006). Engineered antibody Fc variants with enhanced effector function. Proceedings of the National academy of Sciences of the United States of America, 103(11), 4005–4010. https://doi.org/10.1073/pnas.0508123103.
Hwang, I., & Park, S. (2008). Computational design of protein therapeutics. Drug Discovery Today: Technologies, 5(2–3), e43–e48. https://doi.org/10.1016/j.ddtec.2008.11.004.
Caravella, J., & Lugovskoy, A. (2010). Design of next-generation protein therapeutics. Current Opinion in Chemical Biology, 14(4), 520–528. https://doi.org/10.1016/j.cbpa.2010.06.175.
Marshall, S. A., Lazar, G. A., Chirino, A. J., & Desjarlais, J. R. (2003). Rational design and engineering of therapeutic proteins. Drug Discovery Today, 8(5), 212–221. https://doi.org/10.1016/S1359-6446(03)02610-2.
Ravn, U., Didelot, G., Venet, S., Ng, K. T., Gueneau, F., Rousseau, F., et al. (2013). Deep sequencing of phage display libraries to support antibody discovery. Methods, 60(1), 99–110. https://doi.org/10.1016/j.ymeth.2013.03.001.
Hoen, P. A. C., Jirka, S. M. G., TenBroeke, B. R., Schultes, E. A., Aguilera, B., Pang, K. H., et al. (2012). Phage display screening without repetitious selection rounds. Analytical Biochemistry, 421(2), 622–631. https://doi.org/10.1016/j.ab.2011.11.005.
Matochko, W. L., Chu, K., Jin, B., Lee, S. W., Whitesides, G. M., & Derda, R. (2012). Deep sequencing analysis of phage libraries using Illumina platform. Methods, 58(1), 47–55. https://doi.org/10.1016/j.ymeth.2012.07.006.
Barreto, K., Maruthachalam, B. V., Hill, W., Hogan, D., Sutherland, A. R., & Kusalik, A. (2019). Next-generation sequencing-guided identification and reconstruction of antibody CDR combinations from phage selection outputs. Nucleic Acids Research, 5, 2. https://doi.org/10.1093/nar/gkz131.
Wiedmann, M., Wilson, W. J., Czajka, J., Luo, J., Barany, F., & Batt, C. A. (1994). Ligase chain reaction (LCR): Overview and applications. PCR Methods and Applications, 3(4), S51–S64.
Au, L. C., Yang, F. Y., Yang, W. J., Lo, S. H., & Kao, C. F. (1998). Gene synthesis by a LCR-based approach: High-level production of leptin-L54 using synthetic gene in Escherichia coli. Biochemical and Biophysical Research Communications, 248(1), 200–203. https://doi.org/10.1006/bbrc.1998.8929.
Dillon, P. J., & Rosen, C. A. (1990). A rapid method for the construction of synthetic genes using the polymerase chain reaction. BioTechniques, 9(3), 298–300.
Sandhu, G. S., Aleff, R. A., & Kline, B. C. (1992). Dual asymmetric PCR: One-step construction of synthetic genes. BioTechniques, 12(1), 14–16.
Wu, G., Wolf, J. B., Ibrahim, A. F., Vadasz, S., Gunasinghe, M., & Freeland, S. J. (2006). Simplified gene synthesis: A one-step approach to PCR-based gene construction. Journal of Biotechnology, 124(3), 496–503. https://doi.org/10.1016/j.jbiotec.2006.01.015.
Gao, X., Gulari, E., & Zhou, X. (2004). In situ synthesis of oligonucleotide microarrays. Biopolymers, 73(5), 579–596. https://doi.org/10.1002/bip.20005.
Richmond, K. E., Li, M.-H., Rodesch, M. J., Patel, M., Lowe, A. M., Kim, C., et al. (2004). Amplification and assembly of chip-eluted DNA (AACED): A method for high-throughput gene synthesis. Nucleic Acids Research, 32(17), 5011–5018. https://doi.org/10.1093/nar/gkh793.
Moorcroft, M. J., Meuleman, W. R. A., Latham, S. G., Nicholls, T. J., Egeland, R. D., & Southern, E. M. (2005). In situ oligonucleotide synthesis on poly(dimethylsiloxane): A flexible substrate for microarray fabrication. Nucleic Acids Research, 33(8), e75. https://doi.org/10.1093/nar/gni075.
Cox, J. C., Lape, J., Sayed, M. A., & Hellinga, H. W. (2007). Protein fabrication automation. Protein Science, 16(3), 379–390. https://doi.org/10.1110/ps.062591607.
Kunkel, T. A. (1985). Rapid and efficient site-specific mutagenesis without phenotypic selection. Proceedings of the National Academy of Sciences USA, 82(2), 488–492.
de Wildt, R. M. T., Tomlinson, I. M., Mundy, C. R., & Gorick, B. D. (2000). Antibody arrays for high-throughput screening of antibody–antigen interactions. Nature Biotechnology, 18(9), 989–994. https://doi.org/10.1038/79494.
Krebs, B., Rauchenberger, R., Reiffert, S., Rothe, C., Tesar, M., Thomassen, E., et al. (2001). High-throughput generation and engineering of recombinant human antibodies. Journal of Immunological Methods, 254(1–2), 67–84. https://doi.org/10.1016/S0022-1759(01)00398-2.
Gupta, P. B., Onder, T. T., Jiang, G., Tao, K., Kuperwasser, C., Weinberg, R. A., et al. (2009). Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell, 138(4), 645–659. https://doi.org/10.1016/j.cell.2009.06.034.
Bleicher, K. H., Böhm, H.-J., Müller, K., & Alanine, A. I. (2003). A guide to drug discovery: Hit and lead generation: Beyond high-throughput screening. Nature Reviews Drug Discovery, 2(5), 369–378. https://doi.org/10.1038/nrd1086.
Dietrich, J. A., McKee, A. E., & Keasling, J. D. (2010). High-throughput metabolic engineering: Advances in small-molecule screening and selection. Annual Review of Biochemistry, 79(1), 563–590. https://doi.org/10.1146/annurev-biochem-062608-095938.
Fellouse, F. A., Esaki, K., Birtalan, S., Raptis, D., Cancasci, V. J., Koide, A., et al. (2007). High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. Journal of Molecular Biology, 373(4), 924–940. https://doi.org/10.1016/j.jmb.2007.08.005.
Persson, H., Ye, W., Wernimont, A., Adams, J. J., Koide, A., Koide, S., et al. (2013). CDR-H3 diversity is not required for antigen recognition by synthetic antibodies. Journal of Molecular Biology, 425(4), 803–811. https://doi.org/10.1016/j.jmb.2012.11.037.
Tonikian, R., Zhang, Y., Boone, C., & Sidhu, S. S. (2007). Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nature Protocols, 2(6), 1368–1386. https://doi.org/10.1038/nprot.2007.151.
Lee, C. V., Sidhu, S. S., & Fuh, G. (2004). Bivalent antibody phage display mimics natural immunoglobulin. Journal of Immunological Methods, 284(1–2), 119–132. https://doi.org/10.1016/j.jim.2003.11.001.
Baneyx, F. (1999). Recombinant protein expression in Escherichia coli. Current Opinion in Biotechnology, 10(5), 411–421. https://doi.org/10.1016/S0958-1669(99)00003-8.
Hardjasa, A., Ling, M., Ma, K., & Yu, H. (2010). Investigating the effects of DMSO on PCR fidelity using a restriction digest-based method. Journal of Experimental Microbiology and Immunology (JEMI), 14(April), 161–164.
Chevet, E., Lemaie, G., & Katinka, M. D. (1995). Low concentrations of tetramethylammonium chloride increase yield and specificity of PCR. Nucleic Acids Research, 23(16), 3343–3344. https://doi.org/10.1093/nar/23.16.3343.
Chung, C. T., & Miller, R. H. (1988). A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Research, 16(8), 3580. https://doi.org/10.1093/nar/16.8.3580.
Howard, G., & Kaser, M. (2007). Making and using antibodies: A practical handbook. Lavoisier.Fr. Retrieved from http://books.google.com/books?hl=en&lr=&id=yZQiab1lxEYC&oi=fnd&pg=PA1&dq=Making+and+using+antibodies:+A+practical+handbook&ots=5T0RVvlbOw&sig=N0or4QYBKnaQZCpPU7WgKA9Y7Gg.
Slupphaug, G., Eftedal, I., Kavli, B., Bharati, S., Helle, N. M., & Haug, T. (1995). Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry, 34(1), 128–138.
Cedergren-Zeppezauer, E. S., Larsson, G., Olof Nyman, P., Dauter, Z., & Wilson, K. S. (1992). Crystal structure of a dUTPase. Nature, 355(6362), 740–743. https://doi.org/10.1038/355740a0.
Soltes, G., Barker, H., Marmai, K., Pun, E., Yuen, A., & Wiersma, E. J. (2003). A new helper phage and phagemid vector system improves viral display of antibody Fab fragments and avoids propagation of insert-less virions. Journal of Immunological Methods, 274(1–2), 233–244. https://doi.org/10.1016/S0022-1759(02)00294-6.
Soltes, G., Hust, M., Ng, K. K. Y., Bansal, A., Field, J., Stewart, D. I. H., et al. (2007). On the influence of vector design on antibody phage display. Journal of Biotechnology, 127(4), 626–637. https://doi.org/10.1016/j.jbiotec.2006.08.015.
Huang, H., Economopoulos, N. O., Liu, B. A., Uetrecht, A., Gu, J., Jarvik, N., et al. (2015). Selection of recombinant anti-SH3 domain antibodies by high-throughput phage display. Protein Science, 24, 1890–1900. https://doi.org/10.1002/pro.2799.
Sidhu, S. S. (2001). Engineering M13 for phage display. Biomolecular Engineering, 18(2), 57–63. https://doi.org/10.1016/S1389-0344(01)00087-9.
Wals, K., & Ovaa, H. (2014). Unnatural amino acid incorporation in E. coli: current and future applications in the design of therapeutic proteins. Frontiers in Chemistry, 2, 15. https://doi.org/10.3389/fchem.2014.00015.
Chen, G., Gorelik, L., Simon, K. J., Pavlenco, A., Cheung, A., & Brickelmaier, M. (2015). Synthetic antibodies and peptides recognizing progressive multifocal leukoencephalopathyspecific point mutations in polyomavirus JC capsid viral protein 1. mAbs, 7(4), 681–692. https://doi.org/10.1080/19420862.2015.1038447.
Dobson, C. M. (2003). Protein folding and misfolding. Nature, 426(6968), 884–890. https://doi.org/10.1038/nature02261.
Soto, C. (2003). Unfolding the role of protein misfolding in neurodegenerative diseases. Nature Reviews Neuroscience, 4(1), 49–60. https://doi.org/10.1038/nrn1007.
Kiefhaber, T., Rudolph, R., Kohler, H.-H., & Buchner, J. (1991). Protein aggregation in vitro and in vivo: A quantitative model of the kinetic competition between folding and aggregation. Bio/Technology, 9(9), 825–829. https://doi.org/10.1038/nbt0991-825.
Acknowledgements
We would like to thank Y. Cabrera for optimization assistance with the ELISA detection screen and are also grateful to L. Moffat for helpful discussions and comments. This work was supported by the Charles H. Best Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gallo, E. A High-Throughput Platform for the Generation of Synthetic Ab Clones by Single-Strand Site-Directed Mutagenesis. Mol Biotechnol 61, 410–420 (2019). https://doi.org/10.1007/s12033-019-00171-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-019-00171-9