Abstract
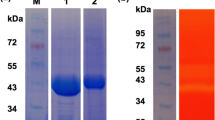
Coptotermes curvignathus is a termite that, owing to its ability to digest living trees, serves as a gold mine for robust industrial enzymes. This unique characteristic reflects the presence of very efficient hydrolytic enzyme systems including cellulases. Transcriptomic analyses of the gut of C. curvignathus revealed that carbohydrate-active enzymes (CAZy) were encoded by 3254 transcripts and that included 69 transcripts encoding glycoside hydrolase family 7 (GHF7) enzymes. Since GHF7 enzymes are useful to the biomass conversion industry, a gene encoding for a GHF7 enzyme (Gh1254) was synthesized, sub-cloned and expressed in the methylotrophic yeast Pichia pastoris. Expressed GH1254 had an apparent molecular mass of 42 kDa, but purification was hampered by its low expression levels in shaken flasks. To obtain more of the enzyme, GH1254 was produced in a bioreactor that resulted in a fourfold increase in crude enzyme levels. The purified enzyme was active towards soluble synthetic substrates such as 4-methylumbelliferyl-β-d-cellobioside, 4-nitrophenyl-β-d-cellobioside and 4-nitrophenyl-β-d-lactoside but was non-hydrolytic towards Avicel or carboxymethyl cellulose. GH1254 catalyzed optimally at 35 °C and maintained 70% of its activity at 25 °C. This enzyme is thus potentially useful in food industries employing low-temperature conditions.








Similar content being viewed by others
Abbreviations
- CMC:
-
Carboxymethyl cellulose
- MUC:
-
4-methylumbelliferyl β-d-cellobioside
- pNPC:
-
p-nitrophenol-β-d-cellobioside
- pNPG3:
-
p-nitrophenol-β-d-cellotrioside
- pNPL:
-
p-nitrophenol-β-d-lactoside
References
Kudo, T. (2009). Termite-microbe symbiotic system and its efficient degradation of lignocellulose. Bioscience, Biotechnology and Biochemistry, 73, 2561–2567.
Ohkuma, M. (2003). Termite symbiotic systems: Efficient bio-recycling of lignocellulose. Applied Microbiology and Biotechnology, 61, 1–9.
US DOE. (2007). Biofuels: Bringing biological solutions to energy challenges. US Department of Energy Office of Science. http://genomicscience.energy.gov/pubs/Biofuels_Flyer_2007-2.pdf.
King, P. J. H., Mahadi, N. M., Bong, C. F. J., Ong, K. H., & Hassan, O. (2014). Bacterial microbiome of Coptotermes curvignathus (Isoptera: Rhinotermitidae) reflects, the coevolution of species and dietary pattern. Insect Science, 21, 584–596.
Brune, A. (1998). Termite guts: The world’s smallest bioreactors. Trends in Biotechnology, 16, 16–21.
Brune, A. (2014). Symbiotic digestion of lignocellulose in termite guts. Nature Reviews, 12, 168–180.
Ni, J., & Tokuda, G. (2013). Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnology Advances, 31, 838–850.
Lo, N., & Eggleton, P. (2011). Termite phylogenetics and co-cladogenesis with symbionts. In D. E. Bignell, Y. Roisin, & N. Lo (Eds.), Biology of termites: A modern synthesis (pp. 27–50). Berlin: Springer.
Tho, Y. P. (1992). Termites of Peninsular Malaysia in Malayan Forest Records No. 36. (Kirton, L.G. ed.). Forest Research Institute Malaysia, Kuala Lumpur, 36, 1–224.
Woon, J. S. K., Mackeen, M. M., Sudin, A. H., Mahadi, N. M., Ilias, R. M., Abdul Murad, A. M., et al. (2016). Production of an oligosaccharide-specific cellobiohydrolase from the thermophilic fungus Thielavia terrestris. Biotechnology Letters, 38(5), 825–832.
Mansfield, S. D., Mooney, C., & Saddler, J. N. (1999). Substrate and enzyme characteristics that limit cellulose hydrolysis. Biotechnology Progress, 15, 804–816.
Woon, J. S. K., Mackeen, M. M., Mahadi, N. M., Ilias, R. M., Abdul Murad, A. M., & Abu Bakar, F. D. (2016). Expression and characterization of a cellobiohydrolase (CBH7B) from the thermophilic fungus Thielavia terrestris in Pichia pastoris. Biotechnology and Applied Biochemistry, 63(5), 690–698.
Teeri, T. T. (1997). Crystalline cellulose degradation: New insight into the function of cellobiohydrolases. Trends in Biotechnology, 15, 160–167.
Nakashima, K., Watanabe, H., & Azuma, J. I. (2002). Cellulase genes from the parabasalian symbiont Pseudotrichonympha grassii in the hindgut of the wood-feeding termite Coptotermes formosanus. Cellular and Molecular Life Sciences, 59, 1554–1560.
Tokuda, G., & Watanabe, H. (2007). Hidden cellulases in termites: Revision of an old hypothesis. Biology Letters, 3, 336–339.
Sethi, A., Kovaleva, E. S., Slack, J. M., Brown, S., Buchman, G. W., & Scharf, M. E. (2013). A GHF7 cellulase from the protist symbiont community of Reticulitermes flavipes enables more efficient lignocellulose processing by host enzymes. Archives of Insect Biochemistry and Physiology, 84(4), 175–193.
Den Haan, R., Mcbride, J. E., Grange, D. C. La, Lynd, L. R., & Van Zyl, W. H. (2007). Functional expression of cellobiohydrolases in Saccharomyces cerevisiae towards one-step conversion of cellulose to ethanol. Enzyme and Microbial Technology, 40, 1291–1299.
Wang, G., Zhang, X., Wang, L., Wang, K., Peng, F., & Wang, L. (2012). The activity and kinetic properties of cellulases in substrates containing metal ions and acid radicals. Advances in Biological Chemistry, 2(11), 390–395.
Cregg, J. M., Vedvick, T. S., & Raschke, W. C. (1993). Recent advances in the expression of foreign genes in Pichia pastoris. Bio-Technology, 11, 905–910.
Valencia, J. A., Wang, H., & Siegfried, B. D. (2014). Expression and characterization of a recombinant endoglucanase from western corn rootworm, in Pichia pastoris. Journal of Insect Science, 1(14), 242.
Sambrook, J. W., & Russell, D. (2001). Molecular cloning: A laboratory manual. New York: Cold Spring Harb Lab Press Cold Spring Harb.
Schulz, M. H., Zerbino, D. R., Vingron, M., & Birney, E. (2012). Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics, 28(8), 1086–1092.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25(17), 3389–3402.
Gilkes, N. R., Henrissat, B., Kilburn, D. G., Miller, R. C., Jr., & Warren, R. A. (1991). Domains in microbial beta-1, 4-glycanases: Sequence conservation, function, and enzyme families. Microbiological Reviews, 55(2), 303–315.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
Lõoke, M., Kristjuhan, K., & Kristjuhan, A. (2011). Extraction of genomic DNA from yeast for PCR-based applications. Biotechniques, 50, 325–328.
Wan Seman, W. M. K., Bakar, S. A., Bukhari, N. A., Gaspar, S. M., Othman, R., Nathan, S., et al. (2014). High level expression of Glomerella cingulata cutinase in dense cultures of Pichia pastoris grown under fed-batch conditions. Journal of Biotechnology, 184, 219–228.
Bradford, M. (1976). Rapid and sensitive method for quantification of microgram quantities of protein utilizing principle of protein-dye-binding. Analytical Biochemistry, 72, 248–254.
Miller, G. L. (1959). Use of dinitrosalicylic reagent for determination of reducing sugar. Analytical Chemistry, 31, 426–428.
Lineweaver, H., & Burk, D. (1934). The determination of enzyme dissociation constants. Journal of the American Chemical Society, 56(3), 658–666.
Hoshino, E., Shiroishi, M., Amano, Y., Nomura, M., & Kanda, T. (1997). Synergistic actions of exo-type cellulases in the hydrolysis of cellulose with different crystallinities. Journal of Fermentation and Bioengineering, 84, 300–306.
Demain, A. L., & Vaishnav, P. (2009). Production of recombinant proteins by microbes and higher organisms. Biotechnology Advances, 27(3), 297–306.
Tomme, P., & Claeyssens, M. (1989). Identification of a functionally important carboxyl group in cellobiohydrolase I from Trichoderma reesei: A chemical modification study. FEBS Letter, 243(2), 239–243.
Humphrey, A. (1998). Shake flask to fermentor: What have we learned? Biotechnology Progress, 14(1), 3–7.
Kyomuhendo, P., Myrnes, B., & Nilsen, I. W. (2007). A cold-active salmon goose-type lysozyme with high heat tolerance. Cellular and Molecular Life Sciences, 64, 2841–2847.
Tokuda, G., Watanabe, H., Matsumoto, T., & Noda, H. (1997). Cellulose digestion in the wood-eating higher termite, Nasutitermes takasagoensis (Shiraki): distribution of cellulases and properties of endo-beta-1,4-glucanase. Zoological Science, 14(1), 83–97.
Inoue, T., Moriya, S., Ohkuma, M., & Kudo, T. (2005). Molecular cloning and characterization of a cellulase gene from a symbiotic protist of the lower termite, Coptotermes formosanus. Gene, 349, 67–75.
Ni, J., Tokuda, G., Takehara, M., & Watanabe, H. (2007). Heterologous expression and enzymatic characterization of β-glucosidase from the drywood-eating termite, Neotermes koshunensis. Applied Entomology and Zoology, 42(3), 457–463.
Evans, T. A., Forschler, B. T., & Grace, J. K. (2013). Biology of invasive termites: A worldwide review. Annual Review of Entomology, 58, 455–474.
Rouland, C., Lenoir-Rousseaux, J. J., Mora, P., & Renoux, J. (1989). Origin of the exocellulase and the beta-glucosidase purified from the digestive tract of the fungus-growing termite Macrotermes muelleri. Sociobiology, 15(1989), 237–246.
McEwen, S. E., Slaytor, M., & O’Brien, R. W. (1980). Cellobiase activity in three species of Australian termites. Insect Biochemistry, 10(5), 563–567.
Ma, R. J., Wang, C. Y., Liu, Y. W., Sivakumar, T. R., Ren, Z. X., Fang, Y., et al. (2014). Identification and characterization of a novel endoglucanase (CMCase) isolated from the larval gut of Bombyx mori. Journal of Asia-Pacific Entomology, 17(1), 67–71.
Beukes, N., & Pletschke, B. I. (2006). Effect of sulfur-containing compounds on Bacillus cellulosome-associated “CMCase” and “Avicelase” activities. FEMS Microbiology Letters, 264(2), 226–231.
Boer, H., Teeri, T. T., & Koivula, A. (2000). Characterization of Trichoderma reesei cellobiohydrolase Cel7a secreted from Pichia pastoris using two different promoters. Biotechnology and Bioengineering, 69, 486–494.
Akcapinar, G. B., Venturini, A., Martelli, P. L., Casadio, R., & Sezerman, U. O. (2015). Modulating the thermostability of Endoglucanase I from Trichoderma reesei using computational approaches. Protein Engineering, Design & Selection, 28(5), 127–135.
Godbole, S., Decker, S. R., Nieves, R. A., Adney, W. S., Vinzant, T. B., Baker, J. O., et al. (1999). Cloning and expression of Trichoderma reesei cellobiohydrolase I in Pichia pastoris. Biotechnological Progress, 15(5), 828–833.
Soares, J. F., Dal Prá, V., Kempka, A. P., Prestes, R. C., Tres, M. V., Kuhn, R. C., et al. (2016). Cellulases for food applications. In V. Gupta (Ed.), New and future developments in microbial biotechnology and bioengineering: Microbial cellulase system properties and applications (pp. 201–208). Amsterdam: Elsevier.
Acknowledgements
The authors would like to thank the Ministry of Science, Technology and Innovation (MOSTI) of Malaysia for providing the research grant 02-05-20-SF11118 and Shaman M. Gaspar (Infors South East Asia/Bumi Sains Sdn. Bhd.) for his help with the bioreactor.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Woon, J.SK., King, P.J.H., Mackeen, M.M. et al. Cloning, Production and Characterization of a Glycoside Hydrolase Family 7 Enzyme from the Gut Microbiota of the Termite Coptotermes curvignathus . Mol Biotechnol 59, 271–283 (2017). https://doi.org/10.1007/s12033-017-0015-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-017-0015-x