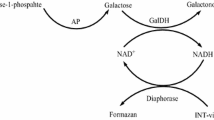
Abstract
Dihydrolipohyl dehydrogenase (DLD) is a FAD-dependent enzyme that catalyzes the reversible oxidation of dihydrolipoamide. Herein, we report medium optimization for the production of a recombinant DLD with NADH-dependent diaphorase activity from a strain of Bacillus sphaericus PAD-91. The DLD gene that consisted of 1413 bp was expressed in Escherichia coli BL21 (DE3), and its enzymatic properties were studied. The composition of production medium was optimized using one-variable-at-a-time method followed by response surface methodology (RSM). B. sphaericus DLD catalyzed the reduction of lipoamide by NAD+ and exhibited diaphorase activity. The molecular weight of enzyme was about 50 kDa and determined to be a monomeric protein. Recombinant diaphorase showed its optimal activity at temperature of 30 °C and pH 8.5. K m and V max values with NADH were estimated to be 0.025 mM and 275.8 U/mL, respectively. Recombinant enzyme was optimally produced in fermentation medium containing 10 g/L sucrose, 25 g/L yeast extract, 5 g/L NaCl and 0.25 g/L MgSO4. At these concentrations, the actual diaphorase activity was calculated to be 345.0 ± 4.1 U/mL. By scaling up fermentation from flask to bioreactor, enzyme activity was increased to 486.3 ± 5.5 U/mL. Briefly, a DLD with diaphorase activity from a newly isolated B. sphaericus PAD-91 was characterized and the production of recombinant enzyme was optimized using RSM technique.










Similar content being viewed by others
References
Liang-Jun, Y., Napporn, T., Nathalie, S., & Michael, J. F. (2013). Serum dihydrolipoamide dehydrogenase is a labile enzyme. Journal of Biochemical and Pharmacological Research, 1, 30–42.
Rachael, A. V., Pierre, R., & Grazia, I. (2011). Mutation in the dimer interface of dihydrolipoamide dehydrogenase promote site-specific oxidative damages in yeast and human cells. The Journal of Biological Chemistry, 286, 40232–40245.
Serrano, A. (1992). Purification, characterization and function of dihydrolipoamide dehydrogenase from cyanobacterium Anabaena sp. strain P.C.C.7119. Biochemistry, 288, 823–830.
Jacques, A. E. B., Willem, J. H. V. B., Walter, M. A. M. V. D., Franz, M., & Arie, D. K. (1989). Molecular cloning and sequence determination of the Ipd gene encoding lipoamide dehydrogenase from Pseudomonas flurescens. Journal of Generral Microbiology, 135, 1787–1797.
Madiagan, R. A., & Mayhew, S. G. (1993). Preparation of the apoenzyme of the FMN-dependent Clostridium kluyveri diaphorase by extraction with apofalvodoxin. Biochemical Society Transactions, 22, 578–585.
Tedeschi, G., Chen, S., & Massey, V. (1995). Active site studies of DT-diaphorase employing artificial flavins. The Journal of Biological Chemistry, 270, 2512–2516.
Melanie, A. A., & Zongchao, J. (2006). Modulator of drug activity B from Escherichia coli: Crystal structure of a prokaryotic homologue of DT-diaphorase. Journal of Molecular Biology, 359, 455–465.
Natalia, L. R., Valentina, A. S., Alexander, V. E., Seergey, V. K., Irina, G. G., Bruce, S. K., et al. (2006). pH-dependent substrate preference of pig heart lipoamide dehydrogenase varies with oligomeric state. The Journal of Biological Chemistry, 280, 16106–16114.
Cakraborty, S., Sakka, M., Kimura, T., & Sakka, K. (2008). Characterization of a dihydrolipoyl dehydrogenase having diaphorase activity of Clostridium kluyveri. Bioscience, Biotechnology, and Biochemistry, 72, 982–988.
Antiochia, R., Cass, A. E. G., & Palleschi, G. (1997). Purification and sensor applications of an oxygen insensitive thermophilic diaphorase. Analytica Chimica Acta, 345, 17–28.
Bhushan, H., & Spain, H. (2002). Diaphorase catalyzed biotransformation of RDX via N-denitration mechanism. Biochemical and Biophysical Research Communications, 296, 779–784.
Shahbazmohammadi, H., & Omidinia, E. (2011). New enzymatic colorimetric method for the quantitative determination of phenylalanine in dry-blood spots. Journal of Sciences, 22, 15–20.
Kianmehr, A., Mahrooz, A., Ansari, J., Oladnabi, M., & Shahbazmohammadi, H. (2016). The rapid and sensitive quantitative determination of galactose by combined enzymatic and colorimetric method: Application in neonatal screening. Applied Biochemistry and Biotechnology, 179, 283–293.
Zhen, H., Liu, Q., & Shao, Y. W. (2009). Medium optimization for the production of a novel bioflocculant from Halomonas sp. V3a using response surface methodology. Bioresource Technology, 100, 5922–5927.
Zhang, H.-B., Mao, X.-Q., Wang, Y.-J., & Hu, X.-Q. (2009). Optimization of culture conditions for high-level expression of dextransucrase in Escherichia coli. The Journal of Food, Agriculture & Environment, 7, 75–78.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1994). Molecular cloning: A laboratory manual (2nd ed., pp. 1847–1857). Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
Shahbazmohammadi, H., & Omidinia, E. (2012). Isolation, purification and characterization of proline dehydrogenase from a newly isolated Pseudomonas putida POS-F84. Iranian Journal of Biotechnology, 10, 111–119.
Argyrou, A., Sun, G., Palfey, B. A., & Blanchard, J. S. (2003). Catalysis of diaphorase reactions by Mycobacterium tuberculosis lipoamide dehydrogenase occurs at the EH4 level. Biochemistry, 42, 2218–2228.
Boething, R. S., & Weaver, T. (1979). A new assay for diaphorase activity in reagent formulations, based on the reduction of thiazolyl blue. Clinical Chemistry, 25, 2040–2042.
Bradford, M. M. (1976). Rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principles of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Ki, J.-S., Zhang, W., & Qian, P.-Y. (2009). Discovery of marine Bacillus species by 16S rRNA and rpoB comparisons and their usefulness for species identification. Journal of Microbiological Methods, 77, 48–57.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739.
Shahbazmohammadi, H., Mostafavi, S. S., Soleimani, S., Bozorgian, S., Pooraskari, M., & Kianmehr, A. (2015). Response surface methodology to optimize partition and purification of two recombinant oxidoreductase enzymes, glucose dehydrogenase and D-galactose dehydrogenase in aqueous two-phase systems. Protein Expression and Purification, 108, 41–47.
Hasan-Beikdashti, M., Forootanfar, H., Safiarian, M. S., Ameri, A., Ghahremani, M. H., Khoshayand, M. R., et al. (2012). Optimization of culture conditions for production of lipase by a newly isolated bacterium Stenotrophomonas maltophilia. Journal of the Taiwan Institute of Chemical Engineers, 43, 670–677.
Rajendran, A., & Thangavelu, V. (2009). Statistical experimental design for evaluation of medium components for lipase production by Rhizopus arrhizus MTCC 2233 LWT-Food. Science Technology, 42, 985–992.
Gupta, N., Mehra, G., & Gupta, R. (2004). A glycerol-inducible lipase from Bacillus sp.: Medium optimization by a Plackett-Burman design and by response surface methodology. Canadian Journal of Microbiology, 50, 361–368.
Singh, J., Vohra, R. M., & Sahoo, D. K. (2004). Enhanced production of alkaline proteases by Bacillus sphaericus using fed-batch culture. Process Biochemistry, 39, 1093–1101.
Gupta, N., Sahai, V., & Gupta, R. (2007). Alkaline lipase from a novel strain Burkholderia multivorans: Statistical medium optimization and production in a bioreactor. Process Biochemistry, 42, 518–526.
Yongxian, F., Yang, Y., Xiaoqin, J., Xiaolong, C., & Yinchu, S. (2013). Cloning, expression and medium optimization of validamycin glycosyltransferase from Streptomyces hygroscopicus var. jinggangensis for the biotransformation of validoxylamine A to produce validamycin A using free resting cells. Bioresource Technology, 131, 13–20.
Dinarvand, M., Rezaee, M., Masomian, M., Jazayeri, S. D., Zareian, M., Abbasi, S., et al. (2013). Effect of C/N Ratio and media optimization through response surface methodology on simultaneous productions of intra and extracellular inulinase and invertase from Aspergillus niger ATCC 20611. BioMed Research International, 2013, 1–13.
Ruchi, G., Anshu, G., & Khare, S. K. (2008). Lipase from solvent tolerant Pseudomonas aeruginosa strain: Production optimization by response surface methodology and application. Bioresource Technology, 99, 4796–4802.
Singh, S., Moholkar, V. S., & Goyal, A. (2014). Optimization of carboxymethylcellulase production from Bacillus amyloliquefaciens SS35. 3 Biotech, 4, 411–424.
Sifour, M., Zaghloul, T. I., Saeed, H. M., Berekaa, M. M., & Abdel-Fattah, Y. R. (2010). Enhanced production of lipase by the thermophilic Geobacillus stearothermophilus strain-5 using statistical experimental designs. New Biotechnology, 27, 330–336.
Acknowledgments
Research finding was from Department of Biochemistry.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Statement
This article does not contain any studies with human participants performed by any of the authors.
Human and Animal Rights Statement
All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
Rights and permissions
About this article
Cite this article
Shahbazmohammadi, H., Omidinia, E. Medium Optimization for Improved Production of Dihydrolipohyl Dehydrogenase from Bacillus sphaericus PAD-91 in Escherichia coli . Mol Biotechnol 59, 260–270 (2017). https://doi.org/10.1007/s12033-017-0013-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-017-0013-z