Abstract
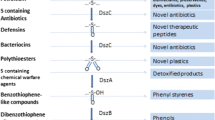
Metabolic pathways of aerobic bacteria able to assimilate sulfur can provide biocatalysts for biodesulfurization of petroleum and of other sulfur-containing pollutants. Of major interest is the so-called “4S pathway,” in that C–S bonds are specifically cleaved leaving the carbon skeleton of substrates intact. This pathway is carried out by four enzymes, named Dsz A, B, C, and D. In view of a possible application of recombinant Dsz enzymes in biodesulfurization treatments, we have investigated the structural features of enzymes cloned from a Rhodococcus strain isolated from polluted environmental samples and their resistance to temperature (20–95 °C) and to organic solvents (5, 10, and 20 % v/v methanol, acetonitrile, hexane, and toluene). Changes in protein structures were assessed by circular dichroism and intrinsic fluorescence spectroscopy. We found that all Dsz proteins are unfolded by temperatures in the range 45–60 °C and by all solvents tested, with the most dramatic effect being produced by toluene. These results suggest that stabilization of the biocatalysts by protein engineering will be necessary for developing biodesulfurization technologies based on Dsz enzymes.




Similar content being viewed by others
Abbreviations
- DBT:
-
Dibenzothiophene
- DBTO:
-
DBT sulfoxide
- DBTO2 :
-
DBT sulfone
- HDS:
-
Hydrodesulfurization
- 2-HBP:
-
2-Hydroxybiphenyl
- FMN:
-
Flavin mononucleotide
References
Mohebali, G., & Ball, A. S. (2008). Biocatalytic desulfurization (BDS) of petrodiesel fuels. Microbiology, 154, 2169–2183.
Babich, I. V., & Moulijn, A. C. (2003). Science and technology of novel processes for deep desulfurization of oil refinery streams: a review. Fuel, 82, 607–631.
Furimsky, E., & Massoth, F. E. (1999). Deactivation of hydroprocessing catalysts. Catalysis Today, 52, 381–495.
McFarland, B. L. (1999). Biodesulfurization. Current Opinion in Microbiology, 2, 257–264.
Kilbane, J. J, 2nd. (2006). Microbial biocatalyst developments to upgrade fossil fuels. Current Opinion in Biotechnology, 17, 305–314.
Li, M. Z., Squires, C. H., Monticello, D. J., & Childs, J. D. (1996). Genetic analysis of the dsz promoter and associated regulatory regions of Rhodococcus erythropolis IGTS8. Journal of Bacteriology, 178, 6409–6418.
Shavandi, M., Soheili, M., Zareian, S., Akbari, N., & Khajeh, K. (2013). The gene cloning, overexpression, purification, and characterization of dibenzothiophenemonooxygenase and desulfinase from Gordonia alkanivorans RIPI90A. Journal of Petroleum Science and Technology, 3, 57–64.
Ma, T., Li, S., Li, G., Wang, R., Liang, F., et al. (2006). Cloning and expressing DBT (dibenzothiophene) monooxygenase gene (dszC) from Rhodococcus sp. DS-3 in Escherichia coli. Frontiers of Biology in China, 4, 375–380.
Matsubara, T., Ohshiro, T., Nishina, Y., & Izumi, Y. (2001). Purification, characterization, and overexpression of flavin reductase involved in dibenzothiophene desulfurization by Rhodococcus erythropolis D-1. Applied and Environment Microbiology, 67, 1179–1184.
Zhang, Q., Tong, M. Y., Li, Y. S., Gao, H. J., & Fang, X. C. (2007). Extensive desulfurization of diesel by Rhodococcus erythropolis. Biotechnology Letters, 29, 123–127.
Yu, B., Ma, C., Zhou, W., Wang, Y., Cai, X., et al. (2006). Microbial desulfurization of gasoline by free whole-cells of Rhodococcus erythropolis XP. FEMS Microbiology Letters, 258, 284–289.
Alves, L., Salgueiro, R., Rodrigues, C., Mesquita, E., Matos, J., et al. (2005). Desulfurization of dibenzothiophene, benzothiophene, and other thiophene analogs by a newly isolated bacterium, Gordonia alkanivorans strain 1B. Applied Biochemistry and Biotechnology, 120, 199–208.
Alves, L., Melo, M., Mendonça, D., Simões, F., Matos, J., Tenreiro, R., & Gírio, F. M. (2007). Sequencing, cloning and expression of the dsz genes required for dibenzothiophene sulfone desulfurization from Gordonia alkanivorans strain 1B. Enzyme and Microbial Technol, 40, 1598–1603.
Tao, F., Yu, B., Xu, P., & Ma, C. Q. (2006). Biodesulfurization in biphasic systems containing organic solvents. Applied and Environment Microbiology, 72, 4604–4609.
Konishi, J., Onaka, T., Ishii, Y., & Suzuki, M. (2000). a) Demonstration of the carbon-sulfur bond targeted desulfurization of benzothiophene by thermophilic Paenibacillus sp. strain A11-2 capable of desulfurizing dibenzothiophene. FEMS Microbiology Letters, 187, 151–154.
Li, G. Q., Ma, T., Li, S. S., Li, H., Liang, F. L., et al. (2007). Improvement of dibenzothiophene desulfurization activity by removing the gene overlap in the dsz operon. Bioscience, Biotechnology, and Biochemistry, 71, 849–854.
Hirasawa, K., Ishii, Y., Kobayashi, M., Koizumi, K., & Maruhashi, K. (2001). Improvement of desulfurization activity in Rhodococcus erythropolis KA2-5-1 by genetic engineering. Bioscience, Biotechnology, and Biochemistry, 65, 239–246.
Studier, F. W. (2005). Protein production by auto-induction in high density shaking cultures. Protein Expression and Purification, 41, 207–234.
Chang, J. H., Rhee, S. K., Chang, Y. K., & Chang, H. N. (1998). Desulfurization of diesel oils by a newly isolated dibenzothiophene-degrading Nocardia sp. strain CYKS2. Biotechnology Progress, 14, 851–855.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning. A laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Sreerama, N., & Woody, R. W. (2000). Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Analytical Biochemistry, 287, 252–260.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402.
Arnold, K., Bordoli, L., Kopp, J., & Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics, 22, 195–201.
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research, 42, W252–W258.
Remmert, M., Biegert, A., Hauser, A., & Soding, J. (2012). HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nature Methods, 9, 173–175.
Piddington, C. S., Kovacevich, B. R., & Rambosek, J. (1995). Sequence and molecular characterization of a DNA region encoding the dibenzothiophene desulfurization operon of Rhodococcus sp. strain IGTS8. Applied and Environment Microbiology, 61, 468–475.
Denome, S. A., Oldfield, C., Nash, L. J., & Young, K. D. (1994). Characterization of the desulfurization genes from Rhodococcus sp. strain IGTS8. Journal of Bacteriology, 176, 6707–6716.
Ellis, H. R. (2010). The FMN-dependent two-component monooxygenase systems. Archives of Biochemistry and Biophysics, 497, 1–12.
Tu, S. C., Lei, B., Liu, M., Tang, C. K., & Jeffers, C. (2000). Probing the mechanisms of the biological intermolecular transfer of reduced flavin. Journal of Nutrition, 130, 331S–332S.
Tu, S. C. (2001). Reduced flavin: donor and acceptor enzymes and mechanisms of channeling. Antioxidants & Redox Signaling, 3, 881–897.
Tu, S. C. (2008). Activity coupling and complex formation between bacterial luciferase and flavin reductases. Photochemical & Photobiological Sciences, 7, 183–188.
Lei, B., & Tu, S. C. (1998). Mechanism of reduced flavin transfer from Vibrio harveyi NADPH-FMN oxidoreductase to luciferase. Biochemistry, 37, 14623–14629.
Jeffers, C. E., & Tu, S. C. (2001). Differential transfers of reduced flavin cofactor and product by bacterial flavin reductase to luciferase. Biochemistry, 40, 1749–1754.
Ohshiro, T., Aoi, Y., Torii, K., & Izumi, Y. (2002). Flavin reductase coupling with two monooxygenases involved in dibenzothiophene desulfurization: purification and characterization from a non-desulfurizing bacterium, Paenibacillus polymyxa A-1. Applied Microbiology and Biotechnology, 59, 649–657.
Louie, T. M., Xie, X. S., & Xun, L. (2003). Coordinated production and utilization of FADH2 by NAD(P)H-flavin oxidoreductase and 4-hydroxyphenylacetate 3-monooxygenase. Biochemistry, 42, 7509–7517.
Gisi, M. R., & Xun, L. (2003). Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:flavin adenine dinucleotide oxidoreductase (TftC) of Burkholderia cepacia AC1100. Journal of Bacteriology, 185, 2786–2792.
Ishii, Y., Konishi, J., Suzuki, M., & Maruhashi, K. (2000). Cloning and expression of the gene encoding the thermophilic NAD(P)H-FMN oxidoreductase coupling with the desulfurization enzymes from Paenibacillus sp. A11-2. Journal of Bioscience and Bioengineering, 90, 591–599.
van Berkel, W. J., Kamerbeek, N. M., & Fraaije, M. W. (2006). Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. Journal of Biotechnology, 124, 670–689.
Li, L., Liu, X., Yang, W., Xu, F., Wang, W., et al. (2008). Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase. Journal of Molecular Biology, 376, 453–465.
Ohshiro, T., Kojima, T., Torii, K., Kawasoe, H., & Izumi, Y. (1999). Purification and characterization of dibenzothiophene (DBT) sulfone monooxygenase, an enzyme involved in DBT desulfurization, from Rhodococcus erythropolis D-1. Journal of Bioscience and Bioengineering, 88, 610–616.
Feng, L., Wang, W., Cheng, J., Ren, Y., Zhao, G., et al. (2007). Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc Natl Acad Sci U S A, 104, 5602–5607.
Liu, S., Zhang, C., Su, T., Wei, T., Zhu, D., et al. (2014). Crystal structure of DszC from Rhodococcus sp. XP at 1.79 A. Proteins, 82, 1708–1720.
Duan, X., Zhang, L., Zhou, D., Ji, K., Ma, T., et al. (2013). Crystallization and preliminary structural analysis of dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis. Acta Crystallographica, Section F: Structural Biology and Crystallization Communications, 69, 597–601.
Zhang, L., Duan, X., Zhou, D., Dong, Z., Ji, K., et al. (2014). Structural insights into the stabilization of active, tetrameric DszC by its C-terminus. Proteins, 82, 2733–2743.
Guan, L. J., Lee, W. C., Wang, S., Ohshiro, T., Izumi, Y. et al. (2015). Crystal structures of apo-DszC and FMN-bound DszC from Rhodococcus erythropolis D-1. FEBS Journal, 282, 3126–3135.
Ohshiro, T., Ohkita, R., Takikawa, T., Manabe, M., Lee, W. C., et al. (2007). Improvement of 2′-hydroxybiphenyl-2-sulfinate desulfinase, an enzyme involved in the dibenzothiophene desulfurization pathway, from Rhodococcus erythropolis KA2-5-1 by site-directed mutagenesis. Bioscience, Biotechnology, and Biochemistry, 71, 2815–2821.
Zhang, Y., Edwards, T. E., Begley, D. W., Abramov, A., Thompkins, K. B., et al. (2011). Structure of nitrilotriacetate monooxygenase component B from Mycobacterium thermoresistibile. Acta Crystallographica, Section F: Structural Biology and Crystallization Communications, 67, 1100–1105.
Knobel, H. R., Egli, T., & van der Meer, J. R. (1996). Cloning and characterization of the genes encoding nitrilotriacetate monooxygenase of Chelatobacter heintzii ATCC 29600. Journal of Bacteriology, 178, 6123–6132.
Holm, L., & Rosenstrom, P. (2010). Dali server: Conservation mapping in 3D. Nucleic Acids Research, 38, W545–W549.
Doble, M., & Kruthiventi, A. K. (2005). Biotreatment of Industrial Effluents. Burlington: Elsevier Butterworth-Heinemann.
Stapleton, R. D. J., & Singh, V. P. E. (2002). Biotransformations: Bioremediation technology for health and environmental protection. In: Progress in industrial microbiology. Amsterdam, The Netherlands: Elsevier B.V.
Kamatari, Y. O., Konno, T., Kataoka, M., & Akasaka, K. (1996). The methanol-induced globular and expanded denatured states of cytochrome c: A study by CD fluorescence, NMR and small-angle X-ray scattering. Journal of Molecular Biology, 259, 512–523.
Uversky, V. N., Narizhneva, N. V., Kirschstein, S. O., Winter, S., & Lober, G. (1997). Conformational transitions provoked by organic solvents in beta-lactoglobulin: can a molten globule like intermediate be induced by the decrease in dielectric constant? Folding and Design, 2, 163–172.
Konishi, J., Ishii, Y., Onaka, T., Ohta, Y., Suzuki, M., et al. (2000). b) Purification and characterization of dibenzothiophene sulfone monooxygenase and FMN-dependent NADH oxidoreductase from the thermophilic bacterium Paenibacillus sp. strain A11-2. Journal of Bioscience and Bioengineering, 90, 607–613.
Bhatia, S., & Sharma, D. K. (2012). Thermophilic desulfurization of dibenzothiophene and different petroleum oils by Klebsiella sp. 13T. Environmental Science and Pollution Research International, 19, 3491–3497.
Aggarwal, S., Karimi, I. A., Kilbane Ii, J. J., & Lee, D. Y. (2012). Roles of sulfite oxidoreductase and sulfite reductase in improving desulfurization by Rhodococcus erythropolis. Molecular BioSystems, 8, 2724–2732.
Santarossa, G., Gatti Lafranconi, P., Alquati, C., DeGioia, L., Alberghina, L., Fantucci, P., & Lotti, M. (2005). Mutations in the ‘‘lid’’ region affect chain length specificity and thermostability of a Pseudomonas fragi lipase. FEBS Letters, 579, 2383–2386.
Acknowledgments
This work was supported by CORIMAV, a consortium between Pirelli and University of Milano-Bicocca, through a doctoral fellowship to F.P. The authors are grateful to C. Santambrogio for fruitful discussion and to J. Pleiss, University of Stuttgart, for help in the bioinformatic analysis and for hosting F.P. for a stage.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interests and that all authors read and approved this manuscript.
Ethical approval
This research did not imply human participants or animals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Parravicini, F., Brocca, S. & Lotti, M. Evaluation of the Conformational Stability of Recombinant Desulfurizing Enzymes from a Newly Isolated Rhodococcus sp.. Mol Biotechnol 58, 1–11 (2016). https://doi.org/10.1007/s12033-015-9897-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-015-9897-7