Abstract
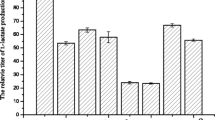
Streptococcus thermophilus is a lactic acid bacterium and used as starter culture in the dairy industry, mainly in the manufacture of yoghurt, with Lactobacillus delbrueckii subsp. bulgaricus. It produces lactic acid as a major fermentation end product and some carbonyl compounds through sugar metabolism. The level of metabolites could be improved using molecular biotechnology. The genes of als, encoding α-acetolactate synthase (Als), the pflA, encoding pyruvate-formate lyase activating enzyme (PflA), and the adhB which encodes alcohol dehydrogenase (AdhB) of S. thermophilus NCFB2393 strain were amplified by polymerase chain reaction and separately cloned into the overexpression vector pNZ276 under the control of the lacA promoter. The strains were transformed individually with the constructed plasmids. Their abilities to generate important metabolites such as pyruvate, lactate, formate, acetaldehyde, acetoin, ethanol, and 2,3-butanediol in LM17 medium were analyzed using high-performance liquid chromatography. High level of 2,3-butanediol was obtained by overexpressing the als gene. The level of formate increased slightly by overexpressing the pflA gene. The overexpression of the adhB gene, on the other hand, resulted in a significant increase in the ethanol level.




Similar content being viewed by others
References
Akyol, I., Serdaroglu, K., Gezginc, Y., Dayisoylu, K. S., Ekinci, M. S., & Ozkose, E. (2009). Redirection of pyruvate pathway of lactic acid bacteria to improve cheese quality. Food Biotechnology, 23, 200–213.
Almiron-Roig, E., Mulholland, F., Gasson, M. J., & Griffin, A. M. (2000). The complete cps gene cluster from Streptococcus thermophilus NCFB 2393 involved in the biosynthesis of a new exopolysaccharide. Microbiol-SGM, 146, 2793–2802.
Arnau, J., Jorgensen, F., Madsen, S. M., Vrang, A., & Israelsen, H. (1997). Cloning, expression, and characterization of the Lactococcus lactis pfl gene, encoding pyruvate formate lyase. Journal of Bacteriology, 179, 5884–5891.
Asanuma, N., Iwamoto, M., & Hino, T. (1999). Structure and transcriptional regulation of the gene encoding pyruvate formate-lyase of a ruminal bacterium, Streptococcus bovis. Microbiology-UK, 145, 151–157.
Asanuma, N., Yoshii, T., & Hino, T. (2004). Molecular characterization of CcpA and involvement of this protein in transcriptional regulation of lactate dehydrogenase and pyruvate formate lyase in the ruminal bacterium Streptococcus bovis. Applied and Environmental Microbiology, 70, 5244–5251.
Beshkova, D. M., Simova, E. D., Frengova, G. I., Simov, Z. I., & Dimitrov, Z. P. (2003). Production of volatile aroma compounds by kefir starter cultures. International Dairy Journal, 13, 529–535.
Bolotin, A., Quinquis, B., Renault, P., Sorokin, A., Ehrlich, S. D., Kulakauskas, S., et al. (2004). Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nature Biotechnology, 22, 1554–1558.
Bongers, R. S., Hoefnagel, M. H. N., & Kleerebezem, M. (2005). High-level acetaldehyde production in Lactococcus lactis by metabolic engineering. Applied and Environmental Microbiology, 71, 1109–1113.
Casadaban, M. J., & Cohen, S. N. (1980). Analysis of gene control signals by DNA-fusion and cloning in Escherichia coli. Journal of Molecular Biology, 138, 179–207.
Chaves, A. C. S. D., Fernandez, M., Lerayer, A. L. S., Mierau, I., Kleerebezem, M., & Hugenholtz, J. (2002). Metabolic engineering of acetaldehyde production by Streptococcus thermophilus. Applied and Environmental Microbiology, 68, 5656–5662.
Courtin, P., & Rul, F. (2004). Interactions between microorganisms in a simple ecosystem: yogurt bacteria as a study model. Lait, 84, 125–134.
de Felipe, F. L., Starrenburg, M. J. C., & Hugenholtz, J. (1997). The role of NADH-oxidation in acetoin and diacetyl production from glucose in Lactococcus lactis subsp. lactis MG1363. FEMS Microbiology Letters, 156, 15–19.
Derzelle, S., Bolotin, A., Mistou, M. Y., & Rul, F. (2005). Proteome analysis of Streptococcus thermophilus grown in milk reveals pyruvate formate lyase as the major upregulated protein. Applied and Environmental Microbiology, 71, 8597–8605.
Gezginc, Y., Topcal, F., Comertpay, S., & Akyol, I. (2015). Quantitative analysis of the lactic acid and acetaldehyde produced by Streptococcus thermophilus and Lactobacillus bulgaricus strains isolated from traditional Turkish yogurts using HPLC. Journal of Dairy Science, 98, 1426–1434.
Guo, X. W., Zhang, Y. H., Cao, C. H., Shen, T., Wu, M. Y., Chen, Y. F., et al. (2014). Enhanced production of 2,3-butanediol by overexpressing acetolactate synthase and acetoin reductase in Klebsiella pneumoniae. Biotechnology and Applied Biochemistry, 61, 707–715.
Hols, P., Hancy, F., Fontaine, L., Grossiord, B., Prozzi, D., Leblond-Bourget, N., et al. (2005). New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiology Reviews, 29, 435–463.
Jensen, N. B. S., Melchiorsen, C. R., Jokumsen, K. V., & Villadsen, J. (2001). Metabolic behavior of Lactococcus lactis MG1363 in microaerobic continuous cultivation at a low dilution rate. Applied and Environmental Microbiology, 67, 2677–2682.
Kang, X., Ling, N., Sun, G., Zhou, Q., Zhang, L., & Sheng, Q. (2012). Complete genome sequence of Streptococcus thermophilus strain MN-ZLW-002. Journal of Bacteriology, 194, 4428–4429.
Karakas-Sen, A., Ridout, M. J., & Narbad, A. (2012). Heterologous expression and purification of the dehydratase NisB involved in the biosynthesis of lantibiotic nisin. Annals of Microbiology, 62, 1099–1107.
Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E., et al. (2006). Comparative genomics of the lactic acid bacteria. Proceedings of the National Academy of Sciences of the United States of America, 103, 15611–15616.
McNulty, N. P., Yatsunenko, T., Hsiao, A., Faith, J. J., Muegge, B. D., Goodman, A. L., et al. (2011). The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Science Translational Medicine, 3, 106.
Melchiorsen, C. R., Jensen, N. B., Christensen, B., Vaever Jokumsen, K., & Villadsen, J. (2001). Dynamics of pyruvate metabolism in Lactococcus lactis. Biotechnology and Bioengineering, 74, 271–279.
Melchiorsen, C. R., Jokumsen, K. V., Villadsen, J., Johnsen, M. G., Israelsen, H., & Arnau, J. (2000). Synthesis and posttranslational regulation of pyruvate formate-lyase in Lactococcus lactis. Journal of Bacteriology, 182, 4783–4788.
Mollet, B., Constable, A., Delley, M., Knol, J., Marciset, O., & Pridmore, D. (1993). Molecular Genetics in Streptococcus thermophilus from transformation to gene-expression. Lait, 73, 175–180.
Ott, A., Germond, J. E., Baumgartner, M., & Chaintreau, A. (1999). Aroma comparisons of traditional and mild yogurts: Headspace gas chromatography quantification of volatiles and origin of alpha-diketones. Journal of Agricultural and Food Chemistry, 47, 2379–2385.
Ott, A., Germond, J. E., & Chaintreau, A. (2000). Origin of acetaldehyde during milk fermentation using C-13-labeled precursors. Journal of Agricultural Food Chemistry, 48, 1512–1517.
Pastink, M. I., Teusink, B., Hols, P., Visser, S., de Vos, W. M., & Hugenholtz, J. (2009). Genome-scale model of Streptococcus thermophilus LMG18311 for metabolic comparison of lactic acid bacteria. Applied and Environmental Microbiology, 75, 3627–3633.
Perez, P. F., Deantoni, G. L., & Anon, M. C. (1991). Formate production by Streptococcus thermophilus cultures. Journal of Dairy Science, 74, 2850–2854.
Sambrook, J., Fritsch, F., & Maniatis, T. (2001). Molecular cloning: a laboratory manual (2nd ed.). Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
Sieuwerts, S., Molenaar, D., van Hijum, S. A. F. T., Beerthuyzen, M., Stevens, M. J. A., Janssen, P. W. M., et al. (2010). Mixed culture transcriptome analysis reveals the molecular basis of mixed culture growth in Streptococcus thermophilus and Lactobacillus bulgaricus. Applied and Environmental Microbiology, 76, 7775–7784.
Sun, Z., Chen, X., Wang, J., Zhao, W., Shao, Y., Wu, L., et al. (2011). Complete genome sequence of Streptococcus thermophilus strain ND03. Journal Bacteriology, 193, 793–794.
Treu, L., Vendramin, V., Bovo, B., Campanaro, S., Corich, V., & Giacomini, A. (2014). Genome Sequences of Streptococcus thermophilus strains MTH17CL396 and M17PTZA496 from Fontina, an Italian PDO cheese. Genome Announcements 2.
Treu, L., Vendramin, V., Bovo, B., Campanaro, S., Corich, V., & Giacomini, A. (2014). Whole-genome sequences of Streptococcus thermophilus strains TH1435 and TH1436, isolated from raw goat milk. Genome Announcements 2.
van den Bogaard, P. T. C., Hols, P., Kuipers, O. P., Kleerebezem, M., & de Vos, W. M. (2004). Sugar utilisation and conservation of the gal-lac gene cluster in Streptococcus thermophilus. Systematic and Applied Microbiology, 27, 10–17.
Vos, P., Vanasseldonk, M., Vanjeveren, F., Siezen, R., Simons, G., & Devos, W. M. (1989). A maturation protein is essential for production of active forms of lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. Journal of Bacteriology, 171, 2795–2802.
Wu, Q., Tun, H. M., Leung, F. C., & Shah, N. P. (2014). Genomic insights into high exopolysaccharide producing dairy starter bacterium Streptococcus thermophilus ASCC 1275. Scientific Reports, 4, 4974.
Zhang, M., Eddy, C., Deanda, K., Finkestein, M., & Picataggio, S. (1995). Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science, 267, 240–243.
Acknowledgments
This work was supported by the Scientific and Technological Research Council of Turkey (TÜBİTAK), Project No: 110 O 218.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Akyol, I., Ozcelik, F.G., Karakas-Sen, A. et al. Cloning and Overexpression of the als, pflA, and adhB Genes in Streptococcus thermophilus and Their Effects on Metabolite Formation. Mol Biotechnol 57, 923–930 (2015). https://doi.org/10.1007/s12033-015-9882-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-015-9882-1