Abstract
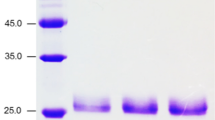
We have previously described two forms of an endo-β-1,4-xylanase (XynSW2A and XynSW2B) synthesized by thermotolerant Streptomyces sp. SWU10. Here, we describe another xylanolytic enzyme, designated XynSW1. The enzyme was purified to homogeneity from 2 L of culture filtrate. Its apparent molecular mass was 24 kDa. The optimal pH and temperature were pH 5.0 and 40 °C, respectively. The enzyme was stable in a wide pH ranges (pH 1–11), more than 80 % of initial activity remained at pH 2–11 after 16 h of incubation at 4 °C and stable up to 50 °C for 1 h. Xylobiose and xylotriose were the major xylooligosaccharides released from oat spelt xylan by the action of XynSW1, indicating of endo-type xylanase. The complete xynSW1 gene contains 1,011 bp in length and encode a polypeptide of 336 with 41 amino acids of signal peptide. The amino acid sequence analysis revealed that it belongs to glycoside hydrolase family 11 (GH11). The mature xynSW1 gene without signal peptide sequence was overexpressed in Pichia pastoris KM71H. The recombinant XynSW1 protein showed higher molecular mass due to the differences in glycosylation levels at the six N-glycosylation sites in the amino acid sequence and exhibited better physicochemical properties than those of the native enzyme including higher optimal temperature (60 °C), and specific activity, but lower optimal pH (4.0). Because of their stability in a wide pH ranges, both of native and recombinant enzymes of XynSW1, may have potential application in several industries including food, textile, biofuel, and also waste treatment.




Similar content being viewed by others
References
Coughlan, M. P., & Hazlewood, G. P. (1993). β-1,4-D-Xylan-degrading enzyme systems: Biochemistry, molecular biology and applications. Biotechnology and Applied Biochemistry, 17, 259–289.
Beg, Q. K., Kapoor, M., Mahajan, L., & Hoondal, G. S. (2001). Microbial xylanases and their industrial applications: A review. Applied Microbiology and Biotechnology, 56, 326–338.
Wong, K. K. Y., Tan, L. U. L., & Saddler, J. N. (1988). Multiplicity of β-1,4-xylanase in microorganisms: Functions and applications. Microbiological Reviews, 52, 305–317.
Henrissat, B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochemical Journal, 280, 309–316.
Henrissat, B., & Bairoch, A. (1993). New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochemical Journal, 293, 781–788.
Subramaniyan, S., & Prema, P. (2002). Biotechnology of microbial xylanases: Enzymology, molecular biology, and application. Critical Reviews in Biotechnology, 22, 33–64.
Wong, K. K. Y., & Saddler, J. N. (1992). Trichoderma xylanases, their properties and application. Critical Reviews in Biotechnology, 12, 413–435.
Butt, M. S., Nadeem, M. T., Ahmad, Z., & Sultan, M. T. (2008). Xylanases and their applications in baking industry. Food Technology and Biotechnology, 46, 22–31.
Bedford, M. R., & Classen, H. L. (1992). Reduction of intestinal viscosity through manipulation of dietary rye and pentosanase concentration is effected through changes in the carbohydrate composition of the intestinal aqueous phase and results in improved growth rate and food conversion efficiency of broiler chicks. Journal of Nutrition, 122, 560–569.
Twomey, D. F., Griffiths, P. C., Hignett, B. C., & Martin, T. P. (2003). Suspected chlamydial polyarthritis in a calf in the UK. Veterinary Record, 152, 340.
Biely, P., Mislovicova, D., & Toman, R. (1985). Soluble chromogenic substrates for the assay of endo-1,4-β-xylanases and endo-1,4-β-glucanases. Analytical Biochemistry, 144, 142–146.
Prade, R. A. (1996). Xylanases: From biology to biotechnology. Biotechnology and Genetic Engineering Reviews, 13, 101–131.
Öhgrena, K., Burab, R., Saddlerb, J., & Zacchia, G. (2007). Effect of hemicellulose and lignin removal on enzymatic hydrolysis of steam pretreated corn stover. Bioresource Technology, 98, 2503–2510.
Chandra, R. P., Au-Yeung, K., Chanis, C., Roos, A. A., Mabee, W., Chung, P. A., et al. (2011). The influence of pretreatment and enzyme loading on the effectiveness of batch and fed-batch hydrolysis of corn stover. Biotechnology Progress, 27, 77–85.
Martín-Sampedro, R., Rodríguez, A., Ferrer, A., García-Fuentevilla, L. L., & Eugenio, M. E. (2012). Biobleaching of pulp from oil palm empty fruit bunches with laccase and xylanase. Bioresource Technology 110,371–378.
Srinivasan, M. C., & Rele, M. V. (1999). Microbial xylanases for paper industry. Current Science, 77, 137–142.
Nelson, N. (1944). A photometric adaption of the Somogyi method for the determination of glucose. Journal of Biological Chemistry, 153, 375–380.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Sakamoto, T., Tsujitani, Y., Fukamachi, K., Taniguchi, Y., & Ihara, H. (2010). Identification of two GH27 bifunctional proteins with β-l-arabinopyranosidase/α-d-galactopyranosidase activities from Fusarium oxysporum. Applied Microbiology and Biotechnology, 86, 1115–1124.
Li, N., Meng, K., Wang, Y., Shi, P., Luo, H., Bai, Y., et al. (2008). Cloning, expression, and characterization of a new xylanase with broad temperature adaptability from Streptomyces sp. S9. Applied Microbiology and Biotechnology, 80, 231–240.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402.
Bendtsen, J. D., Nielsen, H., von Heijne, G., & Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology, 340, 783–795.
Deesukon, W., Nishimura, Y., Harada, N., Sakamoto, T., & Sukhumsirichart, W. (2011). Purification, characterization and gene cloning of two forms of a thermostable endo-xylanase from Streptomyces sp. SWU10. Process Biochemistry, 46, 2255–2262.
Feng, J. X., Karita, S., Fujino, E., Fujino, T., Kimura, T., Sakka, K., et al. (2000). Cloning, sequencing, and expression of the gene encoding a cell-bound multi-domain xylanase from Clostridium josui, and characterization of the translated product. Bioscience, Biotechnology, and Biochemistry, 64, 2614–2624.
Selvaraj, T., Kim, S. K., Kim, Y. H., Jeong, Y. S., Kim, Y. J., Phuong, N. D., et al. (2010). The role of carbohydrate-binding module (CBM) repeat of a multimodular xylanase (XynX) from Clostridium thermocellum in cellulose and xylan binding. Journal of Microbiology, 48, 856–861.
Bentley, S. D., Chater, K. F., Cerdeno-Tarraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature, 417, 141–147.
Brake, A. J., Merryweather, J. P., Coit, D. G., Heberlein, U. A., Masiarz, F. R., Mullenbach, G. T., et al. (1984). α-Factor-directed synthesis and secretion of mature foreign proteins in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America, 81, 4642–4646.
Li, N., Yang, P., Wang, Y., Luo, H., Meng, K., Wu, N., et al. (2008). Cloning, expression, and characterization of protease-resistant xylanase from Streptomyces fradiae var. k11. Journal of Microbiology and Biotechnology, 18, 410–416.
Li, N., Shi, P., Yang, P., Wang, Y., Luo, H., Bai, Y., et al. (2009). A xylanase with high pH stability from Streptomyces sp. S27 and its carbohydrate-binding module with/without linker-region-truncated versions. Applied Microbiology and Biotechnology, 83, 99–107.
Wang, Y., Zhang, H., He, Y., & Luo, H. B. Y. (2007). Characterization, gene cloning, and expression of a novel xylanase XYNB from Streptomyces olivaceoviridis A1. Aquaculture, 267, 328–334.
Li, N., Shi, P., Yang, P., Wang, Y., Luo, H., Bai, Y., et al. (2009). Cloning, expression, and characterization of a new Streptomyces sp. S27 xylanase for which xylobiose is the main hydrolysis product. Applied Biochemistry and Biotechnology, 159, 521–531.
Fu, X. Y., Zhao, W., Xiong, A. S., Tian, Y. S., & Peng, R. H. (2011). High expression of recombinant Streptomyces sp. S38 xylanase in Pichia pastoris by codon optimization and analysis of its biochemical properties. Molecular Biology Reports, 38, 4991–4997.
Sun, J. Y., Liu, M. Q., Weng, X. Y., Qian, L. C., & Gu, S. H. (2007) Expression of recombinant Thermomonospora fusca xylanase A in Pichia pastoris and xylooligosaccharides released from xylans by it. Food Chemistry 104, 1055–1064.
Ninawe, S., Kapoor, M., & Kuhad, R. C. (2008). Purification and characterization of extracellular xylanase from Streptomyces cyaneus SN32. Bioresource Technology, 99, 1252–1258.
Graminha, E. B. N., Gonçalves, A. Z. L., Pirota, R. D. P. B., Balsalobre, M. A. A., & Da Silva, R. E. G. (2008). Enzyme production by solid-state fermentation: Application to animal nutrition. Animal Feed Science and Technology 144, 1–22.
Biely, P., Vrsanska, M., Tenkanen, M., & Kluepfel, D. (1997). Endo-beta-1,4-xylanase families: Differences in catalytic properties. Journal of Biotechnology 57, 151–166.
Biely, P., Vrsanska, M., & Kucar, S. (1992). Identification and mode of action of endo-(1,4)-β-xylanases. In J. Visser, G. Beldman, M. A. Kusters-van-Someren, & A. G. J. Voragen (Eds.), Xylans and xylanases (pp. 81–95). Amsterdam: Elsevier.
Katapodis, P., Vrsansk, M., Kekos, D., Nerinckx, W., Biely, P., Claeyssens, M., et al. (2003). Biochemical and catalytic properties of an endoxylanase purified from the culture filtrate of Sporotrichum thermophile. Carbohydrate Research, 338, 1881–1890.
Vardakou, M., Katapodis, P., Samiotaki, M., Kekos, D., Panayotou, G., & Christakopoulos, P. (2003). Mode of action of family 10 and 11 endoxylanases on water-unextractable arabinoxylan. International Journal of Biological Macromolecules, 33, 129–134.
Pollet, A. (2010). Functional and structural analysis of glycoside hydrolase families (GH) 8, 10 and 11 xylanases with focus on GH11 Bacillus subtilis xylanase A. Ph.D. Thesis, Katholieke Universiteit Leuven, Leuven.
Joshi, M. D., Sidhu, G., Pot, I., Brayer, G. D., Withers, S. G., & McIntosh, L. P. (2000). Hydrogen bonding and catalysis: A novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase. Journal of Molecular Biology, 299, 255–279.
Sapag, A., Wouters, J., Lambert, C., de Ioannes, P., Eyzaguirre, J., & Depiereux, E. (2002). The endoxylanases from family 11: Computer analysis of protein sequences reveal important structural and phylogenetic relationships. Journal of Biotechnology, 95, 109–131.
Krengel, U., & Dijkstra, B. W. (1996). Three-dimensional structure of endo-1,4-betaxylanase I from Aspergillus niger: Molecular basis for its low pH optimum. Journal of Molecular Biology, 263, 70–78.
Fushinobu, S., Ito, K., Konno, M., Wakagi, T., & Matsuzawa, H. (1998). Crystallographic and mutational analyses of an extremely acidophilic and acid-stable xylanase: Biased distribution of acidic residues and importance of Asp37 for catalysis at low pH. Protein Engineering, 11, 1121–1128.
Acknowledgments
This study was supported by grants from the Royal Golden Jubilee Ph. D. Program of Thailand Research Fund (PHD/0257/2549), Srinakharinwirot University (315/2551) and JSPS-NRCT Asian Core Program. The authors thank Dr. Alfredo Villarroel for English proof.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Deesukon, W., Nishimura, Y., Sakamoto, T. et al. Purification, Characterization of GH11 Endo-β-1,4-xylanase from Thermotolerant Streptomyces sp. SWU10 and Overexpression in Pichia pastoris KM71H. Mol Biotechnol 54, 37–46 (2013). https://doi.org/10.1007/s12033-012-9541-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-012-9541-8