Abstract
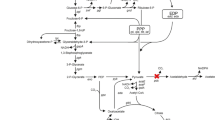
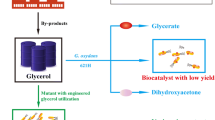
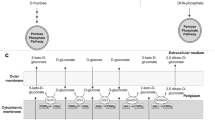
Gluconobacter oxydans is widely used in several biotechnological applications, where sorbitol or mannitol is commonly used as carbon source at high concentration. In this study, a membrane-bound glucose dehydrogenase-deficient strain (GDHK) was constructed to eliminate growth problems on glucose caused by direct oxidation of glucose in the medium. To achieve improved growth properties for the GDHK strain on glucose, a laboratory adaptive evolution experiment was performed with glucose as the sole carbon source. Results indicated evident, albeit modest, improvements in cell growth after a 50-day (about 430 generations) experimental evolution on glucose. The maximum specific growth rate and biomass yield of the resulting GDHE50 strain were increased around 1.35- to 1.4-fold compared with those of the GDHK strain. Meanwhile, two types of biotransformation reactions using resting cells of G. oxydans were investigated. Significant elevations in biotransformation performance of the GHDE50 strain were observed in comparison with that of the wild-type strain. In addition, resting cells of the GDHE50 strain grown on a relatively low concentration of glucose (10 g/l) could catalyze the biotransformation of glycerol to dihydroxyacetone and ethylene glycol to glycolic acid as efficient as the wild-type G. oxydans cultured on higher concentration of sorbitol or other carbon sources. These results suggest very favorable prospects of using glucose to lower production cost in many important industrial biocatalysis and biotransformation processes.




Similar content being viewed by others
References
Deppenmeier, U., & Ehrenreich, A. (2009). Physiology of acetic acid bacteria in light of the genome sequence of Gluconobacter oxydans. Journal of Molecular Microbiology and Biotechnology, 16(1–2), 69–80.
Adachi, O., Moonmangmee, D., Toyama, H., Yamada, M., Shinagawa, E., & Matsushita, K. (2003). New developments in oxidative fermentation. Applied Microbiology and Biotechnology, 60(6), 643–653.
Holscher, T., Schleyer, U., Merfort, M., Bringer-Meyer, S., Gorisch, H., & Sahm, H. (2009). Glucose oxidation and PQQ-dependent dehydrogenases in Gluconobacter oxydans. Journal of Molecular Microbiology and Biotechnology, 16(1–2), 6–13.
Silberbach, M., Maier, B., Zimmermann, M., & Bchs, J. (2003). Glucose oxidation by Gluconobacter oxydans: Characterization in shaking-flasks, scale-up and optimization of the pH profile. Applied Microbiology and Biotechnology, 62(1), 92–98.
Wei, G. D., Yang, X. P., Gan, T. L., Zhou, W. Y., Lin, J. P., & Wei, D. Z. (2009). High cell density fermentation of Gluconobacter oxydans DSM 2003 for glycolic acid production. Journal of Industrial Microbiology & Biotechnology, 36(8), 1029–1034.
Gupta, A., Verma, V., & Qazi, G. (2006). Transposon induced mutation in Gluconobacter oxydans with special reference to its direct-glucose oxidation metabolism1. FEMS Microbiology Letters, 147(2), 181–188.
Gupta, A., Felder, M., Verma, V., Cullum, J., & Qazi, G. (1999). A mutant of Gluconobacter oxydans deficient in gluconic acid dehydrogenase. FEMS Microbiology Letters, 179, 501–506.
Krajewski, V., Simic, P., Mouncey, N. J., Bringer, S., Sahm, H., & Bott, M. (2010). Metabolic engineering of Gluconobacter oxydans for improved growth rate and growth yield on glucose by elimination of gluconate formation. Applied and Environmental Microbiology, 76(13), 4369–4376.
Sambrook, J., Fritsch, E., & Maniatis, T. (1989). Molecular cloning. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Derbise, A., Lesic, B., Dacheux, D., Ghigo, J., & Carniel, E. (2006). A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunology and Medical Microbiology, 38(2), 113–116.
Prust, C., Hoffmeister, M., Liesegang, H., Wiezer, A., Fricke, W. F., Ehrenreich, A., et al. (2005). Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nature Biotechnology, 23(2), 195–200.
Schleyer, U., Bringer-Meyer, S., & Sahm, H. (2008). An easy cloning and expression vector system for Gluconobacter oxydans. International Journal of Food Microbiology, 125(1), 91–95.
Holscher, T., & Gorisch, H. (2006). Knockout and overexpression of pyrroloquinoline quinone biosynthetic genes in Gluconobacter oxydans 621H. Journal of Bacteriology, 188(21), 7668–7676.
Fong, S., Burgard, A., Herring, C., Knight, E., Blattner, F., Maranas, C., et al. (2005). In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnology and Bioengineering, 91(5), 643–648.
Meiberg, J., & Spa, H. (1983). Microbial production of gluconic acid and gluconates. Antonie van Leeuwenhoek, 49(1), 89–90.
Pronk, J., Levering, P., Olijve, W., & Van Dijken, J. (1989). Role of NADP-dependent and quinoprotein glucose dehydrogenases in gluconic acid production by Gluconobacter oxydans. Enzyme and Microbial Technology, 11(3), 160–164.
Kitos, P., Wang, C., Mohler, B., King, T., & Cheldelin, V. (1958). Glucose and gluconate dissimilation in Acetobacter suboxydans. Journal of Biological Chemistry, 233(6), 1295.
Hua, Q., Joyce, A. R., Palsson, B. O., & Fong, S. S. (2007). Metabolic characterization of Escherichia coli strains adapted to growth on lactate. Applied and Environmental Microbiology, 73(14), 4639–4647.
Ameyama, M., Shinagawa, E., Matsushita, K., & ADACHI, O. (1985). Solubilization, purification and properties of membrane-bound glycerol dehydrogenase from Gluconobacter industrius. Agricultural and Biological Chemistry, 49(4), 1001–1010.
Cornish-Bowden, A. (1995). Fundamentals of enzyme kinetics. London: Portland Press.
Krochta, J., Tillin, S., & Hudson, J. (1988). Thermochemical conversion of polysaccharides in concentrated alkali to glycolic acid. Applied Biochemistry and Biotechnology, 17(1), 23–32.
De Ley, J., & Kersters, K. (1964). Oxidation of aliphatic glycols by acetic acid bacteria. Microbiology and Molecular Biology Reviews, 28(2), 164.
Hernandez-Montalvo, V., Martinez, A., Hernandez-Chavez, G., Bolivar, F., Valle, F., & Gosset, G. (2003). Expression of galP and glk in a Escherichia coli PTS mutant restores glucose transport and increases glycolytic flux to fermentation products. Biotechnology and Bioengineering, 83(6), 687–694.
Hua, Q., Joyce, A., Fong, S., & Palsson, B. (2006). Metabolic analysis of adaptive evolution for in silico-designed lactate-producing strains. Biotechnology and Bioengineering, 95(5), 992–1002.
Conrad, T., Joyce, A., Applebee, M., Barrett, C., Xie, B., Gao, Y., et al. (2009). Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biology, 10(10), R118.
Yang, C., Hua, Q., Baba, T., Mori, H., & Shimizu, K. (2003). Analysis of Escherichia coli anaplerotic metabolism and its regulation mechanisms from the metabolic responses to altered dilution rates and phosphoenolpyruvate carboxykinase knockout. Biotechnology and Bioengineering, 84(2), 129–144.
Acknowledgment
This study was supported by Shanghai Pujiang Program (No. 08PJ14038), National Special Fund for State Key Laboratory of Bioreactor Engineering (No. 2060204), SRF for ROCS, SEM, and partially supported by Shanghai Leading Academic Discipline Project (No. B505).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhu, K., Lu, L., Wei, L. et al. Modification and Evolution of Gluconobacter oxydans for Enhanced Growth and Biotransformation Capabilities at Low Glucose Concentration. Mol Biotechnol 49, 56–64 (2011). https://doi.org/10.1007/s12033-011-9378-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9378-6