Abstract
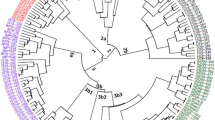
The majority of Hevea (Hevea brasiliensis Muell. Arg.) genetic resource in Vietnam derived from the IRRDB’81 germplasm collected in the Amazonian habitats of the genus. Random amplified polymorphic DNA (RAPD) analysis was used to examine the genetic diversity and structure of the IRRDB’81 germplasm. A total of 59 accessions from 13 different districts of the Brazilian states namely Acre, Rondonia, and Mato Grosso were brought into the study using six arbitrarily preselected primers. Sixty-five RAPD band patterns ranging in size from 0.2 to 3.0 kbp were scored for analysis. Differences in the level of DNA polymorphism among the districts and states were revealed. The percentage of the polymorphic DNA fragments calculated for 13 individual districts varied from 15.38 to 70.77%. The mean values of heterozygosity within the district varied from 0.064 to 0.264. Pairwise district Nei’s genetic distance values ranged from 0.046 for Catriquacu and Itanba of Mato Grosso to 0.304 for Tarauaca of Acre and Aracatuba of Mato Grosso. The estimated values of Shannon’s diversity index ranged from 0.093 for the Assis-Brasil district of Acre to 0.389 for the Jiparana district of Rondonia. The analysis of molecular variance (AMOVA) indicated that most of the genetic variations were found among accessions within the districts, while interdistrict variance component accounted for 14.1% only. The low interdistrict differentiation probably implied an extensive gene flow among them. Both the principal coordinate analysis and UPGMA cluster analysis based on genetic distance values revealed a varying degree of separation among the districts and that conformed to geographical origins of Hevea IRRDB’81 collection.



Similar content being viewed by others
References
Slatkin, M. (1987). Gene flow and the geographic structure of populations. Science, 236, 787–792. doi:10.1126/science.3576198.
Schaal, B. A., Hayworth, D. A., Olsen, K. M., Rauscher, J. T., & Smith, W. A. (1998). Phylogeographic studies in plants: Problems and prospects. Molecular Ecology, 7, 465–474. doi:10.1046/j.1365-294x.1998.00318.x.
Mitton, J. B. (1994). Molecular approaches to population biology. Annual Review of Ecology and Systematics, 25, 45–69. doi:10.1146/annurev.es.25.110194.000401.
Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A., & Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 18, 6531–6535. doi:10.1093/nar/18.22.6531.
Silva, E. P., & Russo, C. A. M. (2000). Techniques and statistical data analysis in molecular population genetics. Hydrobiologia, 420, 119–135. doi:10.1023/A:1003993824352.
Nybom, H. (2004). Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology, 13, 1143–1155. doi:10.1111/j.1365-294X.2004.02141.x.
Volis, S., Yakubov, B., Shulgina, I., Ward, D., Zur, V., & Mendlinger, S. (2001). Tests for adaptive RAPD variation in population genetic structure of wild barley, Hordeum spontaneum Koch. Biological Journal of the Linnean Society. Linnean Society of London, 74, 289–303. doi:10.1006/bijl.2001.0569.
Lynch, M., & Milligan, B. G. (1994). Analysis of population genetic structure with RAPD markers. Molecular Ecology, 3, 91–99. doi:10.1111/j.1365-294X.1994.tb00109.x.
Excoffier, L., Smouse, P. E., & Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics, 131, 479–491.
Huff, D. R., Peakall, R., & Smouse, P. E. (1993). RAPD variation within and among natural populations of outcrossing buffalograss [Buchloë dactyloides (Nutt.) Engelm.]. Theoretical and Applied Genetics, 86, 927–934. doi:10.1007/BF00211043.
Díaz, V., Muniz, L. M., & Ferrer, E. (2001). Random amplified polymorphic DNA and amplified fragment length polymorphism assessment of genetic variation in Nicaraguan populations of Pinus oocarpa. Molecular Ecology, 10, 2593–2603. doi:10.1046/j.0962-1083.2001.01390.x.
Varghese, Y. A., Knaak, C., Sethuraj, M. R., & Ecke, W. (1997). Evaluation of random amplified polymorphic DNA (RAPD) markers in Hevea brasilinesis. Plant Breeding, 116, 47–52. doi:10.1111/j.1439-0523.1997.tb00973.x.
Venkatachalam, P., Thomas, S., Priya, P., Thanseem, I., Saraswathyamma, C. K., Gireesh, T., et al. (2002). Identification of DNA polymorphism among clones of Hevea brasiliensis Muell. Arg. using RAPD analysis. Indian Journal of Natural Rubber Research, 5, 172–181.
Venkatachalam, P., Priya, P., Saraswathyamma, C. K., & Thulaseedharan, A. (2004). Identification, cloning and sequence analysis of a dwarf genome-specific RAPD marker in rubber tree (Hevea brasiliensis Muell. Arg.). Plant Cell Reports, 23, 327–332. doi:10.1007/s00299-004-0833-8.
Tan, H. (1987). Strategies in rubber tree breeding. In A. J. Abbott & R. K. Atkin (Eds.), Improving vegetatively propagated crops (pp. 27–62). London: Academic Press.
Doyle, J. J., & Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry Bulletin, 9, 11–15.
Nei, M. (1972). Genetic distance between populations. American Naturalist, 106, 283–292. doi:10.1086/282771.
Nei, M., & Li, W. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences of the United States of America, 76, 5269–5273. doi:10.1073/pnas.76.10.5269.
Yeh, F. C., Yang, R. C., & Boyle, T. (1999). Popgene: Microsoft window-based freeware for population genetic analysis, version 1.31. Edmonton: University of Alberta.
Peakall, A., & Smouse, P. E. (2006). GenAlEx: Genetic analysis in excel. Population genetic software for teaching and research, version 6. Molecular Ecology Notes, 6, 288–295. doi:10.1111/j.1471-8286.2005.01155.x.
Pavlicek, A., Hrda, S., & Flegr, J. (1999). FreeTree—Freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of the genus Frenkelia. Folia Biologica, 45, 97–99.
Zewei, A., Han, C., Aihua, S., Jialin, F., & Huasun, H. (2005). Identification of rubber clones by RAPD markers. In Proceedings of the International Natural Rubber Conference, 6–8 November, Cochin, India (pp. 88–92).
Venkatachalam, P., Priya, P., Gireesh, T., Saraswathyamma, C. K., & Thulaseedharan, A. (2006). Molecular cloning and sequencing of a polymorphic band from rubber tree (Hevea brasiliensis Muell. Arg.): The nucleotide sequence revealed partial homology with proline-specific permease gene sequence. Current Science, 90, 1510–1515.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583–590.
Shannon, C. E., & Weaver, W. (1949). The mathematical theory of communication. Champaign, IL: University of Illinois Press.
Chevallier, M. H. (1988). Genetic variability of Hevea brasiliensis germplasm using isozyme markers. Journal of Natural Rubber Research, 3, 42–53.
Besse, P., Seguin, M., Lebrun, P., Chevallier, M. H., Nicholas, D., & Lanaud, C. (1994). Genetic diversity among wild and cultivated populations of Hevea brasiliensis assessed by nuclear RFLP analysis. Theoretical and Applied Genetics, 88, 199–207. doi:10.1007/BF00225898.
Lekawipat, N., Teerawatanasuk, K., Rodier-Goud, M., Seguin, M., Vanavichit, A., Toojinda, T., et al. (2003). Genetic diversity analysis of wild germplasm and cultivated clones of Hevea brasiliensis Muell. Arg. by using microsatellite marker. Journal of Rubber Research, 6, 36–47.
Acknowledgments
The research was supported by grant from the Rubber Research Institute of Vietnam. The authors would like to express their gratitude to Mr. Mai Van Son, Director of Rubber Research Institute of Vietnam for his permission to present this paper. Thanks also go to the officers and staffs of Breeding Division of Rubber Research Institute of Vietnam for their contribution to this research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lam, L.V., Thanh, T., Chi, V.T.Q. et al. Genetic Diversity of Hevea IRRDB’81 Collection Assessed by RAPD Markers. Mol Biotechnol 42, 292–298 (2009). https://doi.org/10.1007/s12033-009-9159-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-009-9159-7