Abstract
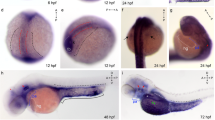
The mechanism by which differentiated cells cooperatively express specific sets of genes in multicellular organisms is a fundamental question for biologists. Currently, the mechanism is primarily attributed to complex regulation of transcriptional machinery. Here, I provide a method for studying spatiotemporal characteristics of promoters in vivo by rapid construction of reporter gene-expression vectors based on simple TA-cloning using an in vivo eGFP reporter assay in Medaka (Oryzias latipes). As an application of this method, I focused on the dopa decarboxylase (Ddc) gene, an essential enzyme for production of neurotransmitters, dopamine, and serotonin. Based on the known structure of the Medaka genome, I predicted and cloned the approximately 3 kbp fragment flanking the Ddc gene. Using an eGFP reporter assay in vivo, I showed that it functions as a promoter, directing reporter gene expression in the brain, retina, epiphysis, and gut, but not in sympathetic ganglia, kidney, or liver. Thus, the procedure presented here provides a useful tool for rapid screening of possible promoter regions and for establishing germ line-transmitted transgenic lines of Medaka.







Similar content being viewed by others
Abbreviations
- Ddc :
-
Dopa decarboxylase
- Hdc :
-
Histamine decarboxylase
- eGFP :
-
Enhanced green fluorescent protein
- MGD:
-
Medaka genome database
- Dpi:
-
Day(s) post injection
- Dpf:
-
Day(s) post fertilization
- UTR:
-
Untranslated region
- Bp:
-
Base pairs
- CNS:
-
Central nervous system
- PNS:
-
Peripheral nervous system
- β-lac:
-
Beta-lactamase
- PCR:
-
Polymerase chain reaction
- HB:
-
Hybridization buffer
References
Wakamatsu, Y., Pristyazhnyuk, S., Kinoshita, M., Tanaka, M., & Ozato, K. (2001). The see-through medaka: A fish model that is transparent throughout life. Proceedings of the National Academy of Sciences of the United States of America, 98, 10046–10050. doi:10.1073/pnas.181204298.
Elsalini, O. A., & Rohr, K. B. (2003). Phenylthiourea disrupts thyroid function in developing zebrafish. Development Genes and Evolution, 212, 593–598.
Wang, W. D., Wang, Y., Wen, H. J., Buhler, D. R., & Hu, C. H. (2004). Phenylthiourea as a weak activator of aryl hydrocarbon receptor inhibiting 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin-induced CYP1A1 transcription in zebrafish embryo. Biochemical Pharmacology, 68, 63–71. doi:10.1016/j.bcp.2004.03.010.
Imai, S., Sasaki, T., Shimizu, A., Asakawa, S., Hori, H., & Shimizu, N. (2007). The genome size evolution of medaka (Oryzias latipes) and fugu (Takifugu rubripes). Genes & Genetic Systems, 82, 135–144. doi:10.1266/ggs.82.135.
Ahsan, B., Kobayashi, D., Yamada, T., Kasahara, M., Sasaki, S., Saito, T. L., et al. (2008). UTGB/medaka: Genomic resource database for medaka biology. Nucleic Acids Research, 36, D747–D752. doi:10.1093/nar/gkm765.
Kasahara, M., Naruse, K., Sasaki, S., Nakatani, Y., Qu, W., Ahsan, B., et al. (2007). The medaka draft genome and insights into vertebrate genome evolution. Nature, 447, 714–719. doi:10.1038/nature05846.
Fujimori, K. E., Kawasaki, T., Deguchi, T., & Yuba, S. (in press). Characterization of a nervous system-specific promoter for growth-associated protein 43 gene in Medaka (Oryzias latipes). Brain Research. doi:10.1016/j.brainres.2008.09.071.
Kumar, S., Tamura, K., & Nei, M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics, 5, 150–163. doi:10.1093/bib/5.2.150.
Borovkov, A. Y., & Rivkin, M. I. (1997). XcmI-containing vector for direct cloning of PCR products. BioTechniques, 22, 812–814.
Goda, N., Tenno, T., Takasu, H., Hiroaki, H., & Shirakawa, M. (2004). The PRESAT-vector: asymmetric T-vector for high-throughput screening of soluble protein domains for structural proteomics. Protein Science, 13, 652–658. doi:10.1110/ps.03439004.
Jo, C., & Jo, S. A. (2001). A simple method to construct T-vectors using XcmI cassettes amplified by nonspecific PCR. Plasmid, 45, 37–40. doi:10.1006/plas.2000.1500.
Chen, Q. J., Zhou, H. M., Chen, J., & Wang, X. C. (2006). Using a modified TA cloning method to create entry clones. Analytical Biochemistry, 358, 120–125. doi:10.1016/j.ab.2006.08.015.
Jeung, J. U., Cho, S. K., Shim, K. S., Ok, S. H., Lim, D. S., & Shin, J. S. (2002). Construction of two pGEM-7Zf(+) phagemid T-tail vectors using AhdI-restriction endonuclease sites for direct cloning of PCR products. Plasmid, 48, 160–163. doi:10.1016/S0147-619X(02)00122-1.
Arashi-Heese, N., Miwa, M., & Shibata, H. (1999). XcmI site-containing vector for direct cloning and in vitro transcription of PCR product. Molecular Biotechnology, 12, 281–283. doi:10.1385/MB:12:3:281.
Jo, C., Kang, B., & Jo, S. A. (2001). Development of new T-vectors containing the luciferase gene. Easy application for direct cloning of a promoter DNA. Molecular Biotechnology, 19, 331–334. doi:10.1385/MB:19:3:331.
Saenz-de-Miera, L. E., & Ayala, F. J. (2004). Complex evolution of orthologous and paralogous decarboxylase genes. Journal of Evolutionary Biology, 17, 55–66. doi:10.1046/j.1420-9101.2003.00652.x.
Ishii, S., Mizuguchi, H., Nishino, J., Hayashi, H., & Kagamiyama, H. (1996). Functionally important residues of aromatic l-amino acid decarboxylase probed by sequence alignment and site-directed mutagenesis. Journal of Biochemistry, 120, 369–376.
Burkhard, P., Dominici, P., Borri-Voltattorni, C., Jansonius, J. N., & Malashkevich, V. N. (2001). Structural insight into Parkinson’s disease treatment from drug-inhibited DOPA decarboxylase. Nature Structural Biology, 8, 963–967. doi:10.1038/nsb1101-963.
Bertoldi, M., Castellani, S., & Bori Voltattorni, C. (2001). Mutation of residues in the coenzyme binding pocket of Dopa decarboxylase. Effects on catalytic properties. European Journal of Biochemistry, 268, 2975–2981. doi:10.1046/j.1432-1327.2001.02187.x.
Ando-Yamamoto, M., Hayashi, H., Sugiyama, T., Fukui, H., Watanabe, T., & Wada, H. (1987). Purification of L-dopa decarboxylase from rat liver and production of polyclonal and monoclonal antibodies against it. Journal of Biochemistry, 101, 405–414.
Shirota, K., & Fujisawa, H. (1988). Purification and characterization of aromatic l-amino acid decarboxylase from rat kidney and monoclonal antibody to the enzyme. Journal of Neurochemistry, 51, 426–434. doi:10.1111/j.1471-4159.1988.tb01056.x.
Kadonaga, J. T. (2004). Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell, 116, 247–257. doi:10.1016/S0092-8674(03)01078-X.
Butler, J. E., & Kadonaga, J. T. (2002). The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes & Development, 16, 2583–2592. doi:10.1101/gad.1026202.
Kleinjan, D. A., & van Heyningen, V. (2005). Long-range control of gene expression: Emerging mechanisms and disruption in disease. American Journal of Human Genetics, 76, 8–32. doi:10.1086/426833.
Forghani, R., Garofalo, L., Foran, D. R., Farhadi, H. F., Lepage, P., Hudson, T. J., et al. (2001). A distal upstream enhancer from the myelin basic protein gene regulates expression in myelin-forming schwann cells. The Journal of Neuroscience, 21, 3780–3787.
Farhadi, H. F., Lepage, P., Forghani, R., Friedman, H. C., Orfali, W., Jasmin, L., et al. (2003). A combinatorial network of evolutionarily conserved myelin basic protein regulatory sequences confers distinct glial-specific phenotypes. The Journal of Neuroscience, 23, 10214–10223.
Taveggia, C., Pizzagalli, A., Fagiani, E., Messing, A., Feltri, M. L., & Wrabetz, L. (2004). Characterization of a Schwann cell enhancer in the myelin basic protein gene. Journal of Neurochemistry, 91, 813–824. doi:10.1111/j.1471-4159.2004.02745.x.
Denarier, E., Forghani, R., Farhadi, H. F., Dib, S., Dionne, N., Friedman, H. C., et al. (2005). Functional organization of a Schwann cell enhancer. The Journal of Neuroscience, 25, 11210–11217. doi:10.1523/JNEUROSCI.2596-05.2005.
Devine-Beach, K., Haas, S., & Khalili, K. (1992). Analysis of the proximal transcriptional element of the myelin basic protein gene. Nucleic Acids Research, 20, 545–550. doi:10.1093/nar/20.3.545.
Hohl, M., & Thiel, G. (2005). Cell type-specific regulation of RE-1 silencing transcription factor (REST) target genes. The European Journal of Neuroscience, 22, 2216–2230. doi:10.1111/j.1460-9568.2005.04404.x.
Chong, J. A., Tapia-Ramirez, J., Kim, S., Toledo-Aral, J. J., Zheng, Y., Boutros, M. C., et al. (1995). REST: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell, 80, 949–957. doi:10.1016/0092-8674(95)90298-8.
Schoenherr, C. J., & Anderson, D. J. (1995). The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science, 267, 1360–1363. doi:10.1126/science.7871435.
Albert, V. R., Lee, M. R., Bolden, A. H., Wurzburger, R. J., & Aguanno, A. (1992). Distinct promoters direct neuronal and nonneuronal expression of rat aromatic l-amino acid decarboxylase. Proceedings of the National Academy of Sciences of the United States of America, 89, 12053–12057. doi:10.1073/pnas.89.24.12053.
Chatelin, S., Wehrle, R., Mercier, P., Morello, D., Sotelo, C., & Weber, M. J. (2001). Neuronal promoter of human aromatic l-amino acid decarboxylase gene directs transgene expression to the adult floor plate and aminergic nuclei induced by the isthmus. Brain Research Molecular Brain Research, 97, 149–160. doi:10.1016/S0169-328X(01)00318-7.
Jahng, J. W., Wessel, T. C., Houpt, T. A., Son, J. H., & Joh, T. H. (1996). Alternate promoters in the rat aromatic l-amino acid decarboxylase gene for neuronal and nonneuronal expression: An in situ hybridization study. Journal of Neurochemistry, 66, 14–19.
Aguanno, A., Lee, M. R., Marden, C. M., Rattray, M., Gault, A., & Albert, V. R. (1995). Analysis of the neuronal promoter of the rat aromatic l-amino acid decarboxylase gene. Journal of Neurochemistry, 65, 1944–1954.
Sumi-Ichinose, C., Hasegawa, S., Ichinose, H., Sawada, H., Kobayashi, K., Sakai, M., et al. (1995). Analysis of the alternative promoters that regulate tissue-specific expression of human aromatic l-amino acid decarboxylase. Journal of Neurochemistry, 64, 514–524.
Visel, A., Thaller, C., & Eichele, G. (2004). GenePaint org: An atlas of gene expression patterns in the mouse embryo. Nucleic Acids Research, 32, D552–D556. doi:10.1093/nar/gkh029.
Odom, D. T., Dowell, R. D., Jacobsen, E. S., Gordon, W., Danford, T. W., MacIsaac, K. D., et al. (2007). Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nature Genetics, 39, 730–732. doi:10.1038/ng2047.
Ason, B., Darnell, D. K., Wittbrodt, B., Berezikov, E., Kloosterman, W. P., Wittbrodt, J., et al. (2006). Differences in vertebrate microRNA expression. Proceedings of the National Academy of Sciences of the United States of America, 103, 14385–14389. doi:10.1073/pnas.0603529103.
Acknowledgment
I would like to thank Yuji Ishikawa for providing Hd-rR and Yuko Wakamatsu for providing SeeThrough via the National BioResource Project (http://www.nbrp.jp/). I also thank Yoriko Jyosaki, Takako Iwanaga, and Kaoru Ogata for excellent assistance in maintaining the fish. This work was supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (#18650095, K.E.F).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fujimori, K.E. Characterization of the Regulatory Region of the Dopa Decarboxylase Gene in Medaka: An in vivo Green Fluorescent Protein Reporter Assay Combined with a Simple TA-Cloning Method. Mol Biotechnol 41, 224–235 (2009). https://doi.org/10.1007/s12033-008-9120-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-008-9120-1