Abstract
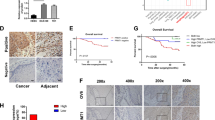
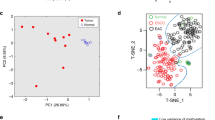
m6A is a widespread RNA modification. However, the mechanism through which m6A regulated the progress of oesophageal squamous cell carcinoma (ESCC) remains undetermined. The levels and prognosis of WTAP were analysed using an ESCC tissue microarray (87 ESCC and 44 paracancerous tissues). TCGA and Oncolnc databases validate WTAP expression and prognosis. CCK8, colony formation (CF), wound healing, transwell cell invasion (CI), and migration (CM) assays were employed for the detection of the biological impacts of WTAP. Expression of tumour stemness-related genes was assessed via qRT-PCR and western blotting. The m6A RNA methylation (m6AMe) quantitative kit was employed for cellular methylation level detection. Arraystar m6A-mRNA and lncRNA epitranscriptomic microarray analyses were used to screen low methylation, high expression, and prognosis-related candidate gene CPSF4. KEGG enrichment analysis was used to screen the downstream signalling pathways of CPSF4. WTAP, a methyltransferase “writer”, was markedly enhanced in ESCC and was strongly correlated with poor patient outcome. WTAP knockdown inhibited the cell proliferation (CP), CI, CM, and stemness of ESCC cells in vitro and reduced the overall m6A modification (m6AMo) percentage of ESCC cells. CPSF4 is a target of WTAP-based m6AMo. WTAP-based m6AMo of CPSF4 transcript reduced the stability of CPSF4 by relying on YTHDF2. We identified the significant role of WTAP-catalysed m6AMo in ESCC tumourigenesis, wherein it facilitates ESCC tumour growth and metastasis through decreasing CPSF4 expression in an m6A-dependent manner.





Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. https://doi.org/10.3322/caac.21660.
Fatehi Hassanabad A, Wong JVS. Statins as potential therapeutics for esophageal cancer. J Gastrointest Cancer. 2021;52:833–8. https://doi.org/10.1007/s12029-021-00648-4.
Sun T, An Q, Yan R, Li K, Zhu K, Dang C, Yuan D. MicroRNA-216a-5p suppresses esophageal squamous cell carcinoma progression by targeting KIAA0101. Oncol Rep. 2020;44:1971–84. https://doi.org/10.3892/or.2020.7751.
Cui Y, Zhang C, Ma S, Li Z, Wang W, Li Y, Ma Y, Fang J, Wang Y, Cao W, Guan F. RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2021;40:294. https://doi.org/10.1186/s13046-021-02096-1.
El Allali A, Elhamraoui Z, Daoud R. Machine learning applications in RNA modification sites prediction. Comput Struct Biotechnol J. 2021;19:5510–24. https://doi.org/10.1016/j.csbj.2021.09.025.
Guo J, Zheng J, Zhang H, Tong J. RNA m6A methylation regulators in ovarian cancer. Cancer Cell Int. 2021;21:609. https://doi.org/10.1186/s12935-021-02318-8.
Qin Y, Li L, Luo E, Hou J, Yan G, Wang D, Qiao Y, Tang C. Role of m6A RNA methylation in cardiovascular disease (Review). Int J Mol Med. 2020;46:1958–72. https://doi.org/10.3892/ijmm.2020.4746.
Ma Z, Li Q, Liu P, Dong W, Zuo Y. METTL3 regulates m6A in endometrioid epithelial ovarian cancer independently of METTl14 and WTAP. Cell Biol Int. 2020;44:2524–31. https://doi.org/10.1002/cbin.11459.
Zheng Q, Yu X, Zhang Q, He Y, Guo W. Genetic characteristics and prognostic implications of m1A regulators in pancreatic cancer. 2021. Biosci Rep. https://doi.org/10.1042/bsr20210337.
Chen M, Wong CM. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol Cancer. 2020;19:44. https://doi.org/10.1186/s12943-020-01172-y.
Li Q, Wang C, Dong W, Su Y, Ma Z. WTAP facilitates progression of endometrial cancer via CAV-1/NF-κB axis. Cell Biol Int. 2021;45:1269–77. https://doi.org/10.1002/cbin.11570.
Chen S, Li Y, Zhi S, Ding Z, Wang W, Peng Y, Huang Y, Zheng R, Yu H, Wang J, Hu M, Miao J, Li J. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m(6)A-dependent manner. Cell Death Dis. 2020;11:659. https://doi.org/10.1038/s41419-020-02847-6.
Chen Y, Peng C, Chen J, Chen D, Yang B, He B, Hu W, Zhang Y, Liu H, Dai L, Xie H, Zhou L, Wu J, Zheng S. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18:127. https://doi.org/10.1186/s12943-019-1053-8.
Yu H, Zhao K, Zeng H, Li Z, Chen K, Zhang Z, Li E, Wu Z. N(6)-methyladenosine (m(6)A) methyltransferase WTAP accelerates the Warburg effect of gastric cancer through regulating HK2 stability. Biomed Pharmacother. 2021;133:111075. https://doi.org/10.1016/j.biopha.2020.111075.
Wei W, Sun J, Zhang H, Xiao X, Huang C, Wang L, Zhong H, Jiang Y, Zhang X, Jiang G. Circ0008399 interaction with WTAP promotes assembly and activity of the m(6)A methyltransferase complex and promotes cisplatin resistance in bladder cancer. Cancer Res. 2021;81:6142–56. https://doi.org/10.1158/0008-5472.CAN-21-1518.
Shriwas O, Mohapatra P, Mohanty S, Dash R. The impact of m6A RNA modification in therapy resistance of cancer: implication in chemotherapy, radiotherapy, and immunotherapy. Front Oncol. 2020;10:612337. https://doi.org/10.3389/fonc.2020.612337.
Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613. https://doi.org/10.1016/j.biopha.2019.108613.
Xu J, Chen Q, Tian K, Liang R, Chen T, Gong A, Mathy NW, Yu T, Chen X. m6A methyltransferase METTL3 maintains colon cancer tumorigenicity by suppressing SOCS2 to promote cell proliferation. Oncol Rep. 2020;44:973–86. https://doi.org/10.3892/or.2020.7665.
Liu X, Xiao M, Zhang L, Li L, Zhu G, Shen E, Lv M, Lu X, Sun Z. The m6A methyltransferase METTL14 inhibits the proliferation, migration, and invasion of gastric cancer by regulating the PI3K/AKT/mTOR signaling pathway. J Clin Lab Anal. 2021;35:e23655. https://doi.org/10.1002/jcla.23655.
Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, Feng Y, Pan Q, Wan R. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. 2020;19:91. https://doi.org/10.1186/s12943-020-01158-w.
Hao L, Wang JM, Liu BQ, Yan J, Li C, Jiang JY, Zhao FY, Qiao HY, Wang HQ. m6A-YTHDF1-mediated TRIM29 upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. Biochim Biophys Acta Mol Cell Res. 2021;1868:118878. https://doi.org/10.1016/j.bbamcr.2020.118878.
Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in cancer progression. Mol Cancer. 2020;19:88. https://doi.org/10.1186/s12943-020-01204-7.
Wang CQ, Tang CH, Wang Y, Huang BF, Hu GN, Wang Q, Shao JK. Upregulated WTAP expression appears to both promote breast cancer growth and inhibit lymph node metastasis. Sci Rep. 2022;12:1023. https://doi.org/10.1038/s41598-022-05035-y.
Zhang M, Lin H, Ge X, Xu Y. Overproduced CPSF4 promotes cell proliferation and invasion via PI3K-AKT signaling pathway in oral squamous cell carcinoma. J Oral Maxillofac Surg. 2021;79:1177.e1-1177.e14. https://doi.org/10.1016/j.joms.2020.12.047.
Chen W, Qin L, Wang S, Li M, Shi D, Tian Y, Wang J, Fu L, Li Z, Guo W, Yu W, Yuan Y, Kang T, Huang W, Deng W. CPSF4 activates telomerase reverse transcriptase and predicts poor prognosis in human lung adenocarcinomas. Mol Oncol. 2014;8:704–16. https://doi.org/10.1016/j.molonc.2014.02.001.
Wang X, Dong J, Li X, Cheng Z, Zhu Q. CPSF4 regulates circRNA formation and microRNA mediated gene silencing in hepatocellular carcinoma. Oncogene. 2021;40:4338–51. https://doi.org/10.1038/s41388-021-01867-6.
Zhang L, Wan Y, Zhang Z, Jiang Y, Lang J, Cheng W, Zhu L. FTO demethylates m6A modifications in HOXB13 mRNA and promotes endometrial cancer metastasis by activating the WNT signalling pathway. RNA Biol. 2021;18:1265–78. https://doi.org/10.1080/15476286.2020.1841458.
Zaccara S, Jaffrey SR. A unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell. 2020;181:1582-1595.e18. https://doi.org/10.1016/j.cell.2020.05.012.
Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, Wong CC, Ng IO, Wong CM. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–70. https://doi.org/10.1002/hep.29683.
Xu F, Li J, Ni M, Cheng J, Zhao H, Wang S, Zhou X, Wu X. FBW7 suppresses ovarian cancer development by targeting the N6-methyladenosine binding protein YTHDF2. Mol Cancer. 2021;20:45. https://doi.org/10.1186/s12943-021-01340-8.
Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, Ma H, Kang T. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–61. https://doi.org/10.1016/j.canlet.2018.11.006.
Funding
This work was supported by the National Natural Science Foundation of China [Grant No. 31870352], 2021 Scientific Research Projects of the Health Commission of Anhui Province [Grant No. AHWJ2021b059], and 2021 Anhui Provincial Postgraduate Scientific Research Project of Universities [Grant No. YJS20210552].
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by QL, XZ, and YK. The first draft of the manuscript was written by QL and XZ and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The organisation chip was approved by Shanghai Outdo Biotech Co., Ltd (Permission No.: SHYJS-CP-1807014).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Luo, Q., Zhan, X., Kuang, Y. et al. WTAP promotes oesophageal squamous cell carcinoma development by decreasing CPSF4 expression in an m6A-dependent manner. Med Oncol 39, 231 (2022). https://doi.org/10.1007/s12032-022-01830-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-022-01830-9