Abstract
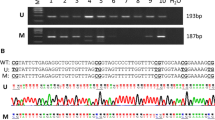
This study is to explore the roles of gene methylation of Notch1 and JAG1 in development of invasive ductal carcinoma of breast. Quantitative analysis the DNA methylation levels of Notch1 and JAG1 gene by the MassARRAY method in invasive ductal carcinoma of breast (IDC; n = 89), atypical ductal hyperplasia of breast (ADH; n = 11), and ordinary ductal hyperplasia of breast (UDH; n = 20). The expressions of JAG1 and Notch1 protein in four breast tissues were detected by immunohistochemistry SP method. (1) Positive expression rates of Notch1 protein in IDC and DCIS were 88.7 % (79/89) and 70.0 % (14/20), respectively, which were significantly higher than the levels in ADH (36.0 %, 4/11) and UDH (25.0 %, 5/20; P < 0.05). Notch1 protein expression was significant positively correlated with lymph node metastasis, pathological grades, and TNM stages of IDC. (2) Positive expression rates of JAG1 protein in IDC and DCIS were 89.9 % (80/89) and 75.0 % (15/20), respectively, which were significantly higher than those of ADH (45.0 %, 5/11) and UDH (30.0 %, 6/20; P < 0.05). JAG1 protein expression was significant positive correlation with lymph node metastasis, pathological grades and TNM stages of IDC. There is an overall hypomethylation alteration of Notch1 and JAG gene in IDC, with corresponding over-expression of Notch1 and JAG1 protein. This inverse correlation shows that the alteration of protein expression results from hypomethylation oncogene Notch1 and JAG1, and this change may play an important role in occurrence and progression of breast cancer.















Similar content being viewed by others
References
Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumor clinicopathological parameters in human breast cancer. Int J Mol Med. 2004;14(5):779–86.
Hao L, Rizzo P, Osipo C, et al. Notch-1 activates estrogen receptor-alpha-dependent transcription via IK Kalpha in breast cancer cells. Oncogene. 2010;29(2):201–13.
Mitsuhashi Y, Horiuchi A, Miyamoto T, et al. Prognostic significance of Notch signaling molecules and their involvement in the invasiveness of endometrial carcinoma cells. Histopathology. 2012;60(5):826–37.
Koch U, Radtke F. Notch signaling in solid tumors. Curr Top Dev Biol. 2010;92:411–55.
Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–59.
Agrawal Anshu, Murphy RF, Agrawal DK. DNA methylation in breast and colorectal cancers. Mod Pathol. 2007;20(7):711–21.
Itoh H, Lwasaki M, Kasuga Y, et al. Association between serum organochlorines and global methylation level of leukocyte DNA among Japanese women: a cross-sectional study. Sci Total Environ. 2014;490(15):603–9.
Siegel R, Ma J, Jemal A. Cancer statistics 2014. CA Cancer J Clin. 2014;64(1):9–29.
Han J, Hendzel MJ, Allalunis TJ. Notch signaling as a therapeutic target for breast cancer treatment? Breast Cancer Res. 2011;13(3):210–6.
Jonusiene V, Sasnauskiene A, Lachei N, et al. Down-regulated expression of Notch signaling molecules in human endometrial cancer. Med Oncol. 2013;30(1):438–45.
Wael H, Yoshida R, Kudoh S, et al. Notch1 signaling controls cell proliferation, apoptosis and differentiation in lung carcinoma. Lung Cancer. 2014;13(5):210–4.
Dominguez M. Oncogenic programmes and Notch activity: an ‘organized crime’? Semin Cell Dev Biol. 2014;28(4):78–85.
Zardwi SJ, Zardawi I, McNeil CM, et al. High Notch1 protein expression is an early event in breast cancer development and is associated with the HER-2 molecular subtype. Histopathology. 2010;56(3):286–96.
Reedijk M, Odorcic S, Chang L, et al. High-level coexpression of JAG1 and notch1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–7.
Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 2014;54(5):716–27.
Ellis L, Atadja PW, Johnstone RW. Epigeneties in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8(6):1409–20.
Haller F, Moskalev EA, Faucz FR, et al. Aberrant DNA hypermethylation of SDHC: a novel mechanism of tumor development in Carney triad. Endocr Relat Cancer. 2014;21(4):567–77.
Gautrey HE, van Otterdijk SD, Cordell HJ, et al. DNA methylation abnormalities at gene promoters are extensive and variable in the elderly and phenocopy cancer cells. FASEB J. 2014;28(7):3261–72.
Avraham Ayelet, Cho Sean Soonweng, Uhlmann Ronit, et al. Tissue specific DNA methylation in normal human breast epithelium and in breast cancer. PLoS One. 2014;9(3):1–8.
Tokaraz P, Blasiak J. Role of DNA methylation in colorectal cancer. Postepy Biochem. 2013;59(3):267–79.
Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14(3):2–4.
Fukushige S, Horii A. DNA methylation in cancer: a gene silencing mechanism and the clinical potential of its biomarkers. Tohoku J Exp Med. 2013;229(3):173–85.
Sharma G, Mirza S, Parshad R, et al. CpG hypomethylation of MDR1 gene in tumor and serum of invasive ductal breast carcinoma patients[J]. Clin Biochem. 2010;43(4–5):373–9.
Fujita N, Kagara N, Yamamoto N, et al. Methylated DNA and high total DNA levels in the serum of patients with breast cancer following neoadjuvant chemotherapy are predictive of a poor prognosis. Oncol Lett. 2014;8(1):397–403.
Rao X, Evans J, Chae H, et al. CpG island shore methylation regulates caveolin-1 expression in breast cancer. Oncogene. 2013;32(38):4519–28.
Jiao F, Bai SY, Ma Y, et al. DNA methylation of heparanase promoter influences its expression and associated with the progression of human breast cancer. PLoS One. 2014;9(3):1–12.
Heyn H, Sayols S, Moutinho C, et al. Linkage of DNA methylation quantitative trait loci to human cancer risk. Cell Rep. 2014;7(2):331–8.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
We declare that we have no financial or commercial conflicts of interest pertaining to this study.
Rights and permissions
About this article
Cite this article
Sun, H., Li, K. & Shen, S. A study of the role of Notch1 and JAG1 gene methylation in development of breast cancer. Med Oncol 33, 35 (2016). https://doi.org/10.1007/s12032-016-0750-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-016-0750-z