Abstract
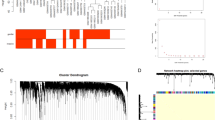
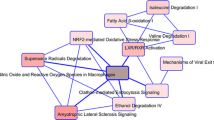
Pituitary adenomas are the most common intrasellar tumors. Patients should be identified at an early stage so that effective treatment can be implemented. The study aims at detecting the potential biomarkers with diagnostic value of pituitary adenomas. Using a total of seven gene expression profiles (GEPs) of the datasets from the Gene Expression Omnibus (GEO) database, we first screened 1980 significant differentially expressed genes (DEGs). Then, we employed the prediction analysis for microarray (PAM) algorithm to identify 340 significant DEGs able to differ pituitary tumor from normal samples, which include 208 upregulated DEGs and 132 downregulated DEGs. DAVID database was used to carry out the enrichment analysis on Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) pathways. We found that upregulated candidates were enriched in protein folding and metabolic pathways. Downregulated DEGs saw a significant enrichment in insulin receptor signaling pathway and hedgehog signaling pathway. Based on the protein-protein interaction (PPI) network as well as module analysis, we determined ten hub genes including PHLPP, ENO2, ACTR1A, EHHADH, EHMT2, FOXO1, DLD, CCT2, CSNK1D, and CETN2 that could be potential biomarkers with diagnostic value in pituitary adenomas. In conclusion, the study contributes to reliable and potential molecular biomarkers with diagnostic value. Moreover, these potential biomarkers may be used for prognosis and new therapeutic targets for the pituitary adenomas.






Similar content being viewed by others
Availability of Data and Materials
The datasets generated during this study are available in the online GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Abbreviations
- BP:
-
Biological process
- CC:
-
Cellular constituent
- DEGs:
-
Differentially expressed genes
- EHHADH:
-
Enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase
- ENO2:
-
Enolase 2
- FC:
-
Fold change
- FDR:
-
False discovery rate
- FOXO1:
-
Forkhead box O1
- GEO:
-
Gene Expression Omnibus
- GEPs:
-
Gene expression profiles
- GO:
-
Gene Ontology
- HCC:
-
Hepatocellular carcinoma cell
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- MF:
-
Molecular function
- PAM:
-
Prediction analysis for microarray
- PHLPP:
-
PH domain leucine-rich repeat protein phosphatase
- PPI:
-
Protein-protein interaction
References
Chu CM, Yao CT, Chang YT, Chou HL, Chou YC, Chen KH, Terng HJ, Huang CS, Lee CC, Su SL, Liu YC, Lin FG, Wetter T, Chang CW (2014) Gene expression profiling of colorectal tumors and normal mucosa by microarrays meta-analysis using prediction analysis of microarray, artificial neural network, classification, and regression trees. Dis Markers 2014:634123. https://doi.org/10.1155/2014/634123
Ezzat S, Asa SL, Couldwell WT, Barr CE, Dodge WE, Vance ML, McCutcheon IE (2004) The prevalence of pituitary adenomas: a systematic review. Cancer 101(3):613–619. https://doi.org/10.1002/cncr.20412
Fan S, Liang Z, Gao Z, Pan Z, Han S, Liu X, Zhao C, Yang W, Pan Z, Feng W (2018) Identification of the key genes and pathways in prostate cancer. Oncol Lett 16(5):6663–6669. https://doi.org/10.3892/ol.2018.9491
Gao X, Wang X, Zhang S (2018) Bioinformatics identification of crucial genes and pathways associated with hepatocellular carcinoma. Biosci Rep 38(6):BSR20181441. https://doi.org/10.1042/BSR20181441
Gueorguiev M, Grossman AB (2011) Pituitary tumors in 2010: a new therapeutic era for pituitary tumors. Nat Rev Endocrinol 7(2):71–73. https://doi.org/10.1038/nrendo.2010.233
Hernandez-Ramirez LC, Morgan RML, Barry S, D’Acquisto F, Prodromou C, Korbonits M (2018) Multi-chaperone function modulation and association with cytoskeletal proteins are key features of the function of AIP in the pituitary gland. Oncotarget 9(10):9177–9198. https://doi.org/10.18632/oncotarget.24183
Huang d W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. https://doi.org/10.1038/nprot.2008.211
Lampron A, Bourdeau I, Hamet P, Tremblay J, Lacroix A (2006) Whole genome expression profiling of glucose-dependent insulinotropic peptide (GIP)- and adrenocorticotropin-dependent adrenal hyperplasias reveals novel targets for the study of GIP-dependent Cushing’s syndrome. J Clin Endocrinol Metab 91(9):3611–3618. https://doi.org/10.1210/jc.2006-0221
Liu ZP, Wu C, Miao H, Wu H (2015) RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database: the journal of biological databases and curation 2015:bav095. https://doi.org/10.1093/database/bav095
Liu CC, Wang H, Wang WD, Wang L, Liu WJ, Wang JH, Geng QR, Lu Y (2018) ENO2 promotes cell proliferation, glycolysis, and glucocorticoid-resistance in acute lymphoblastic leukemia. Cell Physiol Biochem: international journal of experimental cellular physiology, biochemistry, and pharmacology 46(4):1525–1535. https://doi.org/10.1159/000489196
Luo T, Chen X, Zeng S, Guan B, Hu B, Meng Y, Liu F, Wong T, Lu Y, Yun C, Hocher B, Yin L (2018) Bioinformatic identification of key genes and analysis of prognostic values in clear cell renal cell carcinoma. Oncol Lett 16(2):1747–1757. https://doi.org/10.3892/ol.2018.8842
Mathioudakis N, Salvatori R (2009) Pituitary tumors. Curr Treat Options Neurol 11(4):287–296
Michaelis KA, Knox AJ, Xu M, Kiseljak-Vassiliades K, Edwards MG, Geraci M, Kleinschmidt-DeMasters BK, Lillehei KO, Wierman ME (2011) Identification of growth arrest and DNA-damage-inducible gene beta (GADD45beta) as a novel tumor suppressor in pituitary gonadotrope tumors. Endocrinology 152(10):3603–3613. https://doi.org/10.1210/en.2011-0109
Molitch ME (2017) Diagnosis and treatment of pituitary adenomas: a review. Jama 317(5):516–524. https://doi.org/10.1001/jama.2016.19699
Morris DG, Musat M, Czirjak S, Hanzely Z, Lillington DM, Korbonits M, Grossman AB (2005) Differential gene expression in pituitary adenomas by oligonucleotide array analysis. Eur J Endocrinol 153(1):143–151. https://doi.org/10.1530/eje.1.01937
Nijaguna MB, Patil V, Hegde AS, Chandramouli BA, Arivazhagan A, Santosh V, Somasundaram K (2015) An eighteen serum cytokine signature for discriminating glioma from normal healthy individuals. PLoS One 10(9):e0137524. https://doi.org/10.1371/journal.pone.0137524
Qin Y, Meng L, Fu Y, Quan Z, Ma M, Weng M, Zhang Z, Gao C, Shi X, Han K (2017) SNORA74B gene silencing inhibits gallbladder cancer cells by inducing PHLPP and suppressing Akt/mTOR signaling. Oncotarget 8(12):19980–19996. https://doi.org/10.18632/oncotarget.15301
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47. https://doi.org/10.1093/nar/gkv007
Shangkuan WC, Lin HC, Chang YT, Jian CE, Fan HC, Chen KH, Liu YF, Hsu HM, Chou HL, Yao CT, Chu CM, Su SL, Chang CW (2017) Risk analysis of colorectal cancer incidence by gene expression analysis. PeerJ 5:e3003. https://doi.org/10.7717/peerj.3003
Smith AJ, Wen YA, Stevens PD, Liu J, Wang C, Gao T (2016) PHLPP negatively regulates cell motility through inhibition of Akt activity and integrin expression in pancreatic cancer cells. Oncotarget 7(7):7801–7815. https://doi.org/10.18632/oncotarget.6848
Strickland AL, Rivera G, Lucas E, John G, Cuevas I, Castrillon DH (2018) PI3K pathway effectors pAKT and FOXO1 as novel markers of endometrioid intraepithelial neoplasia. Int J Gynecol Pathol: official journal of the International Society of Gynecological Pathologists. https://doi.org/10.1097/PGP.0000000000000549
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45(D1):D362–D368. https://doi.org/10.1093/nar/gkw937
Tibshirani R, Hastie T, Narasimhan B, Chu G (2002) Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A 99(10):6567–6572. https://doi.org/10.1073/pnas.082099299
Tong Y, Zheng Y, Zhou J, Oyesiku NM, Koeffler HP, Melmed S (2012) Genomic characterization of human and rat prolactinomas. Endocrinology 153(8):3679–3691. https://doi.org/10.1210/en.2012-1056
Vierimaa O, Georgitsi M, Lehtonen R, Vahteristo P, Kokko A, Raitila A, Tuppurainen K, Ebeling TM, Salmela PI, Paschke R, Gundogdu S, De Menis E, Makinen MJ, Launonen V, Karhu A, Aaltonen LA (2006) Pituitary adenoma predisposition caused by germline mutations in the AIP gene. Science 312(5777):1228–1230. https://doi.org/10.1126/science.1126100
Wang W, Xu Z, Fu L, Liu W, Li X (2014) Pathogenesis analysis of pituitary adenoma based on gene expression profiling. Oncol Lett 8(6):2423–2430. https://doi.org/10.3892/ol.2014.2613
Xiong X, Wen YA, Mitov MI, M CO, Miyamoto S, Gao T (2017) PHLPP regulates hexokinase 2-dependent glucose metabolism in colon cancer cells. Cell Death Dis 3:16103. https://doi.org/10.1038/cddiscovery.2016.103
Yu Y, Pan X, Ding Y, Liu X, Tang H, Shen C, Shen H, Yang P (2013) An iTRAQ based quantitative proteomic strategy to explore novel secreted proteins in metastatic hepatocellular carcinoma cell lines. Analyst 138(16):4505–4511. https://doi.org/10.1039/c3an00517h
Zhao P, Hu W, Wang H, Yu S, Li C, Bai J, Gui S, Zhang Y (2015) Identification of differentially expressed genes in pituitary adenomas by integrating analysis of microarray data. Int J Endocrinol 2015:164087. https://doi.org/10.1155/2015/164087
Zhou W, Ma CX, Xing YZ, Yan ZY (2016) Identification of candidate target genes of pituitary adenomas based on the DNA microarray. Mol Med Rep 13(3):2182–2186. https://doi.org/10.3892/mmr.2016.4785
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81702643, 81770980, 81570906), China Postdoctoral Science Foundation (Grant No. 2017M611586), Science Project of Shanghai (Grant No. 14DZ2260300), Science Project of Shanghai Municipal Commission of Health and Family Planning (Grant No. 201540173), Shanghai sailing program (Grant No.16YF1403400), and The Second Military Medical University Project (Grant No. 2017JS18).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Not applicable.
Conflict of Interest
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Peng, H., Deng, Y., Wang, L. et al. Identification of Potential Biomarkers with Diagnostic Value in Pituitary Adenomas Using Prediction Analysis for Microarrays Method. J Mol Neurosci 69, 399–410 (2019). https://doi.org/10.1007/s12031-019-01369-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-019-01369-x