Abstract
Purpose
Colorectal cancer (CRC) is one of the most commonly diagnosed malignant tumors and highly heterogeneous diseases. More recently, RNA expression profiles have been used as prognostic cancer markers. In this regard, the expression of small non-coding RNAs like tRNA-derived fragments (tRFs) in tumor tissue has potential diagnostic values in metastatic cancer.
Method
Sixty postoperative CRC tissue samples, consisting of 30 cancers and 30 adjacent normal tissues, were collected from cancer patients. We evaluated MINTbase database to select tRNA-derived fragments. The expression levels of miR-1280, miR1308, tRNA-ValAAC/CAC, and tRNA-AspGTC were measured by TaqMan quantitative reverse transcription PCR technology. Also, we have evaluated the correlation between the levels of tRFs gene expression and clinicopathological of CRC disease.
Result
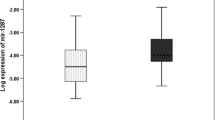
The three tRFs derived from tRF/miR-1280, tRNA-ValAAC/CAC, and tRNA-AspGTC downregulated in tumor tissues (all, p < 0.0001). These tRFs have lower expression in stage IV in comparison with stage III. The tRFs derived from tRNA-ValAAC (p = 0.005) and tRNA-AspGTC (p = 0.034) showed the decreased expression in CRC patients with distant metastasis.
Conclusion
The present study demonstrated that low expression of tRF/miR-1280, tRNA-ValAAC/CAC, and tRNA-AspGTC was significantly associated with metastatic stage and more aggressive tumor behavior of CRC disease. Our finding promising the potential of using tRFs as biomarkers for cancer diagnosis.




Similar content being viewed by others
Availability of Data and Materials
The data and materials that support the findings of this study are available from the corresponding author upon reasonable request.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Chen B, Morgan GJ, Barlogie B, Epstein J. Specific exosomal microRNA are differentially expressed between high and low-risk myeloma suggesting they are pathogenically important. ed: American Society of Hematology Washington, DC 2015.
Balatti V, Pekarsky Y, Croce CM. Role of the tRNA-derived small RNAs in cancer: new potential biomarkers and target for therapy. Adv Cancer Res. 2017;135:173–87.
Grillone K, et al. Non-coding RNAs in cancer: platforms and strategies for investigating the genomic “dark matter.” J Exp Clin Cancer Res. 2020;39(1):1–19.
Zhu L, Ge J, Li T, Shen Y, Guo J. tRNA-derived fragments and tRNA halves: the new players in cancers. Cancer Lett. 2019;452:31–7.
Shan S, Wang Y, Zhu C. A comprehensive expression profile of tRNA‐derived fragments in papillary thyroid cancer. J Clin Lab Anal. 2021;35(3):e23664.
Papadimitriou M-A, et al. tRNA-derived fragments (tRFs) in bladder cancer: increased 5′-tRF-LysCTT results in disease early progression and patients’ poor treatment outcome. Cancers. 2020;12(12):3661.
Ma Z, Zhou J, Shao Y, Jafari FA, Qi P, Li Y. Biochemical properties and progress in cancers of tRNA-derived fragments. J Cell Biochem. 2020;121(3):2058–63.
Huang B, et al. tRF/miR-1280 suppresses stem cell–like cells and metastasis in colorectal cancer. Cancer Res. 2017;77(12):3194–206.
Jones C, et al. Identification of circulating microRNAs as diagnostic biomarkers for use in multiple myeloma. Br J Cancer. 2021;107(12):1987–96.
Li X, et al. Identification of aberrantly expressed miRNAs in rectal cancer. Oncol Rep 2012;28(1):77–84.
Lerebours F, Clairac G, Tozlu-Kara S, Vacher S, Lidereau R, Bieche I. MicroRNA expression profiling of inflammatory breast cancer. ed: AACR.
Yao D, et al. OncotRF: an online resource for exploration of tRNA-derived fragments in human cancers. RNA Biol. 2020;17(8):1081–91.
Zhu L, et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer. 2019;18(1):1–5.
Pliatsika V, et al. MINTbase v2. 0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects. Nucleic Acids Res. 2018;46(D1):D152–9.
Weiser MR. AJCC 8th edition: colorectal cancer. Ann Surg Oncol. 2018;25(6):1454–5.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods, 2001;25(4):402–8.
Sun V, et al. Antitumor activity of miR-1280 in melanoma by regulation of Src. Mol Ther. 2015;23(1):71–8.
Wang F, et al. A microRNA-1280/JAG2 network comprises a novel biological target in high-risk medulloblastoma. Oncotarget. 2015;6(5):2709.
Piepoli A, et al. Mirna expression profiles identify drivers in colorectal and pancreatic cancers. PloS One. 2012;7(3):e33663.
Guo Y, et al. Transfer RNA detection by small RNA deep sequencing and disease association with myelodysplastic syndromes. BMC Genomics. 2015;16(1):1–11.
Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161(4):790–802.
Vafaei S, Fattahi F, Sahlolbei M, Kiani J, Yazdanpanah A, Madjd Z. Dynamic signature of tRNA-derived small RNAs in cancer pathogenesis as a promising valuable approach. Critical Reviews™ in Eukaryotic Gene Expression. 2020;30(5).
Nientiedt M, Deng M, Schmidt D, Perner S, Müller SC, Ellinger J. Identification of aberrant tRNA-halves expression patterns in clear cell renal cell carcinoma. Sci Rep. 2016;6(1):1–9.
Lee K, Ferguson LR. MicroRNA biomarkers predicting risk, initiation and progression of colorectal cancer. World J Gastroenterol. 2016;22(33):7389.
Falzone L et al. Integrated analysis of colorectal cancer microRNA datasets: Identification of microRNAs associated with tumor development. Aging (Albany NY). 2018;10(5):1000.
Li H, et al. Long non-coding RNA 1308 promotes cell invasion by regulating the miR-124/ADAM 15 axis in non-small-cell lung cancer cells. Cancer Manag Res. 2018;10:6599.
Lerebours F, et al. miRNA expression profiling of inflammatory breast cancer identifies a 5‐miRNA signature predictive of breast tumor aggressiveness. Int J Cancer. 2013;133(7):1614–23.
He T, Zheng K, Li G, Song B, Zhang Y. Identification of typical miRNAs and target genes in hepatocellular carcinoma by DNA microarray technique. Eur Rev Med Pharmacol Sci. 2014;18(1):108–16.
Funding
This project was given funds from the Oncopathology Research Center, Iran University of Medical Sciences, Tehran, Iran (grant number 97–4-28–13918).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
This article was conducted under the Ethics Committee of Iran medical university (IR.IUMS.REC.1397.1121). All of the cases and controls gave their informed consent before their inclusion in the study.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sahlolbei, M., Fattahi, F., Vafaei, S. et al. Relationship Between Low Expressions of tRNA-Derived Fragments with Metastatic Behavior of Colorectal Cancer. J Gastrointest Canc 53, 862–869 (2022). https://doi.org/10.1007/s12029-021-00773-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-021-00773-0