Abstract
Purpose
Computational approaches have been used at different stages of drug development with the purpose of decreasing the time and cost of conventional experimental procedures. Lately, techniques mainly developed and applied in the field of artificial intelligence (AI), have been transferred to different application domains such as biomedicine.
Methods
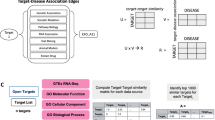
In this study, we conducted an investigative analysis via data-driven evaluation of potential hepatocellular carcinoma (HCC) therapeutics in the context of AI-assisted drug discovery/repurposing. First, we discussed basic concepts, computational approaches, databases, modeling approaches, and featurization techniques in drug discovery/repurposing. In the analysis part, we automatically integrated HCC-related biological entities such as genes/proteins, pathways, phenotypes, drugs/compounds, and other diseases with similar implications, and represented these heterogeneous relationships via a knowledge graph using the CROssBAR system.
Results
Following the system-level evaluation and selection of critical genes/proteins and pathways to target, our deep learning-based drug/compound-target protein interaction predictors DEEPScreen and MDeePred have been employed for predicting new bioactive drugs and compounds for these critical targets. Finally, we embedded ligands of selected HCC-associated proteins which had a significant enrichment with the CROssBAR system into a 2-D space to identify and repurpose small molecule inhibitors as potential drug candidates based on their molecular similarities to known HCC drugs.
Conclusions
We expect that these series of data-driven analyses can be used as a roadmap to propose early-stage potential inhibitors (from database-scale sets of compounds) to both HCC and other complex diseases, which may subsequently be analyzed with more targeted in silico and experimental approaches.




Similar content being viewed by others
Availability of Data and Material
Results/data are available as supplementary data.
Code Availability
All of the utilized computational methods are available as tools/web-services/repositories. CROssBAR: https://crossbar.kansil.org and https://github.com/cansyl/CROssBAR, DEEPScreen: https://github.com/cansyl/DEEPScreen, MDeePred: https://github.com/cansyl/MDeePred, iBioProVis: https://ibpv.kansil.org/.
References
Xue H, et al. Review of drug repositioning approaches and resources. Int J Biol Sci. 2018;14(10):1232–44. https://doi.org/10.7150/ijbs.24612.
Van Norman GA. Drugs, devices, and the FDA: part 1: an overview of approval processes for drugs. JACC Basic Transl Sci. 2016;1(3):170–9. https://doi.org/10.1016/j.jacbts.2016.03.002.
Yeu Y, Yoon Y, Park S. Protein localization vector propagation: a method for improving the accuracy of drug repositioning. Mol Biosyst. 2015;11(7):2096–102. https://doi.org/10.1039/c5mb00306g.
Seyhan AA. Lost in translation: the valley of death across preclinical and clinical divide – identification of problems and overcoming obstacles. Transl Med Commun. 2019;4(1):18. https://doi.org/10.1186/s41231-019-0050-7.
Pushpakom S, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18(1):41–58. https://doi.org/10.1038/nrd.2018.168.
Vamathevan J, et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18(6):463–77. https://doi.org/10.1038/s41573-019-0024-5.
Law V, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42(Database issue):D1091–7. https://doi.org/10.1093/nar/gkt1068.
Rifaioglu AS, et al. Recent applications of deep learning and machine intelligence on in silico drug discovery: methods, tools and databases. Brief Bioinform. 2019;20(5):1878–912. https://doi.org/10.1093/bib/bby061.
Wang Y, et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37(Web Server issue):W623–33. https://doi.org/10.1093/nar/gkp456.
Gaulton A, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40(Database issue):D1100-7. https://doi.org/10.1093/nar/gkr777.
Wishart DS, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–72. https://doi.org/10.1093/nar/gkj067.
Kuhn M, et al. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36(Database issue):D684–8. https://doi.org/10.1093/nar/gkm795.
Gilson MK, et al. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44(D1):D1045–53. https://doi.org/10.1093/nar/gkv1072.
Benson ML, et al. Binding MOAD, a high-quality protein-ligand database. Nucleic Acids Res. 2008;36(Database issue):D674–8. https://doi.org/10.1093/nar/gkm911.
Kanehisa M, et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38(Database issue):D355–60. https://doi.org/10.1093/nar/gkp896.
Kuhn M, et al. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016;44(D1):D1075–9. https://doi.org/10.1093/nar/gkv1075.
Liu Y, et al. DCDB: drug combination database. Bioinformatics. 2010;26(4):587–8. https://doi.org/10.1093/bioinformatics/btp697.
Wishart DS, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35(Database issue):D521–6. https://doi.org/10.1093/nar/gkl923.
Wishart D, et al. T3DB: the toxic exposome database. Nucleic Acids Res. 2015;43(Database issue):D928–34. https://doi.org/10.1093/nar/gku1004.
Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Info Comput Sci. 1988;28(1):31–36. https://doi.org/10.1021/ci00057a005.
Heller SR, et al. InChI, the IUPAC International Chemical Identifier. J Cheminfo. 2015;7(1):23. https://doi.org/10.1186/s13321-015-0068-4.
Dogan T, et al. CROssBAR: comprehensive resource of biomedical relations with knowledge graph representations. Nucleic Acids Res. 2021. https://doi.org/10.1093/nar/gkab543.
Rifaioglu AS, et al. DEEPScreen: high performance drug-target interaction prediction with convolutional neural networks using 2-D structural compound representations. Chem Sci. 2020;11(9):2531–57. https://doi.org/10.1039/c9sc03414e.
Rifaioglu AS, et al. MDeePred: novel multi-channel protein featurization for deep learning-based binding affinity prediction in drug discovery. Bioinformatics. 2021;37(5):693–704. https://doi.org/10.1093/bioinformatics/btaa858.
Wu Y, et al. Cancer stem cells: a potential breakthrough in HCC-targeted therapy. Front Pharmacol. 2020;11:198. https://doi.org/10.3389/fphar.2020.00198.
Nault JC, et al. The role of telomeres and telomerase in cirrhosis and liver cancer. Nat Rev Gastroenterol Hepatol. 2019;16(9):544–58. https://doi.org/10.1038/s41575-019-0165-3.
Manmadhan S, Ehmer U. Hippo signaling in the liver - a long and ever-expanding story. Front Cell Dev Biol. 2019;7:33. https://doi.org/10.3389/fcell.2019.00033.
Ersahin T, Ozturk M, Cetin-Atalay R. Molecular biology of liver cancer, in Rev Cell Biol Mol Med. 2015;206–243.
Wands JR, Kim M. WNT/beta-catenin signaling and hepatocellular carcinoma. Hepatology. 2014;60(2):452–4. https://doi.org/10.1002/hep.27081.
von Olshausen G, et al. Hepatitis B virus promotes beta-catenin-signalling and disassembly of adherens junctions in a Src kinase dependent fashion. Oncotarget. 2018;9(74):33947–33960. https://doi.org/10.18632/oncotarget.26103.
Moon H, Ro SW. MAPK/ERK signaling pathway in hepatocellular carcinoma. Cancers (Basel). 2021;13(12). https://doi.org/10.3390/cancers13123026.
Shang N, et al. Focal adhesion kinase and beta-catenin cooperate to induce hepatocellular carcinoma. Hepatology. 2019;70(5):1631–45. https://doi.org/10.1002/hep.30707.
Vitiello M, et al. Multiple pathways of SIRT6 at the crossroads in the control of longevity, cancer, and cardiovascular diseases. Ageing Res Rev. 2017;35:301–11. https://doi.org/10.1016/j.arr.2016.10.008.
Zha Y, et al. Downregulation of Rap1 promotes 5-fluorouracil-induced apoptosis in hepatocellular carcinoma cell line HepG2. Oncol Rep. 2014;31(4):1691–8. https://doi.org/10.3892/or.2014.3033.
Ngo MT, et al. The role of IGF/IGF-1R signaling in hepatocellular carcinomas: stemness-related properties and drug resistance. Int J Mol Sci. 2021;22(4). https://doi.org/10.3390/ijms22041931.
Marin JJG, et al. Molecular bases of drug resistance in hepatocellular carcinoma. cancers (Basel). 2020;12(6). https://doi.org/10.3390/cancers12061663.
Alzofon N, Jimeno A. Capmatinib for non-small cell lung cancer. Drugs Today (Barc). 2021;57(1):17–25. https://doi.org/10.1358/dot.2021.57.1.3239638.
Qin S, et al. A phase II study of the efficacy and safety of the MET inhibitor capmatinib (INC280) in patients with advanced hepatocellular carcinoma. Ther Adv Med Oncol. 2019;11:1758835919889001. https://doi.org/10.1177/1758835919889001.
Dimri M, Satyanarayana A. Molecular signaling pathways and therapeutic targets in hepatocellular carcinoma. Cancers (Basel). 2020;12(2). https://doi.org/10.3390/cancers12020491.
Fallahi P, et al. Sorafenib and thyroid cancer. BioDrugs. 2013;27(6):615–28. https://doi.org/10.1007/s40259-013-0049-y.
Kane RC, et al. Sorafenib for the treatment of advanced renal cell carcinoma. Clin Cancer Res. 2006;12(24):7271–8. https://doi.org/10.1158/1078-0432.CCR-06-1249.
Llovet JM, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–90. https://doi.org/10.1056/NEJMoa0708857.
Al-Salama ZT, Syed YY, Scott LJ. Lenvatinib: A review in hepatocellular carcinoma. Drugs. 2019;79(6):665–74. https://doi.org/10.1007/s40265-019-01116-x.
Motzer R, et al. Lenvatinib plus pembrolizumab or everolimus for advanced renal cell carcinoma. N Engl J Med. 2021;384(14):1289–300. https://doi.org/10.1056/NEJMoa2035716.
Stjepanovic N, Capdevila J. Multikinase inhibitors in the treatment of thyroid cancer: specific role of lenvatinib. Biologics. 2014;8:129–39. https://doi.org/10.2147/BTT.S39381.
Kuwana M, Azuma A. Nintedanib: new indication for systemic sclerosis-associated interstitial lung disease. Mod Rheumatol. 2020;30(2):225–31. https://doi.org/10.1080/14397595.2019.1696505.
Awasthi N, Schwarz RE. Profile of nintedanib in the treatment of solid tumors: the evidence to date. Onco Targets Ther. 2015;8:3691–701. https://doi.org/10.2147/OTT.S78805.
Yamanaka T, et al. Nintedanib inhibits intrahepatic cholangiocarcinoma aggressiveness via suppression of cytokines extracted from activated cancer-associated fibroblasts. Br J Cancer. 2020;122(7):986–94. https://doi.org/10.1038/s41416-020-0744-7.
Sakurai T, Kudo M. Molecular link between liver fibrosis and hepatocellular carcinoma. Liver Cancer. 2013;2(3–4):365–6. https://doi.org/10.1159/000343851.
Zhang Q, et al. Intratumoral heterogeneity of hepatocellular carcinoma: from single-cell to population-based studies. World J Gastroenterol. 2020;26(26):3720–36. https://doi.org/10.3748/wjg.v26.i26.3720.
Kahraman DC, Kahraman T, Cetin-Atalay R. Targeting PI3K/Akt/mTOR pathway identifies differential expression and functional role of IL8 in liver cancer stem cell enrichment. Mol Cancer Ther. 2019;18(11):2146–57. https://doi.org/10.1158/1535-7163.MCT-19-0004.
Shakiba E, Sadeghi M, Shakiba M. A systematic review and meta-analysis of evaluation of serum interleukin 8 levels in hepatocellular carcinoma. Clin Exp Hepatol. 2019;5(2):123–8. https://doi.org/10.5114/ceh.2019.84780.
Yu LX, Ling Y, Wang HY. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis Oncol. 2018;2(1):6. https://doi.org/10.1038/s41698-018-0048-z.
Li Y, et al. IL-6/STAT3 signaling contributes to sorafenib resistance in hepatocellular carcinoma through targeting cancer stem cells. Onco Targets Ther. 2020;13:9721–30. https://doi.org/10.2147/OTT.S262089.
Donmez A, et al. iBioProVis: interactive visualization and analysis of compound bioactivity space. Bioinformatics. 2020;36(17):4674. https://doi.org/10.1093/bioinformatics/btaa666.
Van der Maaten L, Hinton G. Visualizing data using t-SNE. J Machine Learn Res. 2008;9:2579–2605.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
R. C. A., D. C. K., M. V. A., and T. D. conceived the idea and planned the work. A. S. R., A. A., A. D., H. A., R. C. A., V. A., and T. D. constructed the utilized methods and tools. R. C. A., D. C. K., and E. N. performed the data analysis. R. C. A., D. C. K., E. N., A. C. A., V. A., and T. D. wrote the manuscript. R. C. A., A. C. A., V. A., and T. D. supervised the overall study. All the authors have approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cetin-Atalay, R., Kahraman, D.C., Nalbat, E. et al. Data Centric Molecular Analysis and Evaluation of Hepatocellular Carcinoma Therapeutics Using Machine Intelligence-Based Tools. J Gastrointest Canc 52, 1266–1276 (2021). https://doi.org/10.1007/s12029-021-00768-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-021-00768-x