Abstract
Aim
Gastric cancer (GC) is one of the most common malignant tumors globally, with an increasing incidence rate. Nuclear paraspeckle assembly transcript 1 (NEAT1) is a long non-coding RNA (lncRNAs) responsible for regulating cell cycle progression, apoptosis, cell growth, proliferation, and migration in various cells. The present survey was performed to assess the effects of NEAT1 gene knocking out by CRISPR/Cas9 system in human gastric cancer cells.
Methods
The CRISPR/Cas9 genome editing technique was used to knockout NEAT1 in AGS cells as a gastric cancer model. After the design and construction of the vector, transfection was performed. The expression levels of mRNA, the survival of cells, apoptosis, and cell migration were evaluated by real-time quantitative polymerase chain reaction, flow cytometry, and scratch wound.
Results
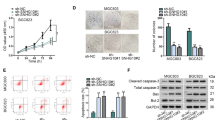
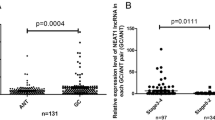
Degradation of NEAT1 by CRISPR/cas9 significantly suppressed the gene’s expression rate, arrested cell cycle in the G0/G1 phase, and a significant reduction in cell number in the S phase (P < 0.05). Degradation of NEAT1 by CRISPR/cas9 also restrained the ability to migrate in transfected cells compared to the control group (P < 0.01). Knockout of NEAT1 via impact on miR-34a gene expression induced apoptosis of AGS cells (P < 0.05) with increasing in the FAS level and total apoptosis (P < 0.001).
Conclusions
Findings suggest that NEAT1 plays a vital role in cellular mechanisms of GC’s occurrence and can serve as a new treatment target in GC.







Similar content being viewed by others
References
Sitarz R, et al. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–48.
Siegel RL, Miller KD, Jemal A. Cancer statistics. Ca Cancer J Clin. 2018;68(1):7–30.
Khodavirdipour A, et al. Apoptosis detection methods in diagnosis of cancer and their potential role in treatment: advantages and disadvantages: a review. J Gastrointest Cancer. 2021.
Desai AG, et al. Medicinal plants and cancer chemoprevention. Curr Drug Metab. 2008;9(7):581–91.
Khodavirdipour A, Zarean R, Safaralizadeh R. Evaluation of the anti-cancer effect of Syzygium cumini ethanolic extract on HT-29 colorectal cell line. J Gastrointest Cancer. 2020:1–7.
Khodavirdipour A, et al. To study in vitro anti-proliferative and pro-apoptotic properties of Salmonella typhi in human pancreatic cancer cell line. Avicenna J Clin Microbiol Infect. 2019;6(3):77–82.
Khodavirdipour A, et al. To study the anti-cancer effects of Shigella flexneri in AspC-1 pancreatic cancer cell line in approach to Bax and bcl-2 genes. J Gastrointest Cancer. 2020:1–7.
Harisa GI, et al. Bacteriosomes as a promising tool in biomedical applications: immunotherapy and drug delivery. AAPS PharmSciTech. 2020;21(5):168.
Farhood B, Geraily G, Alizadeh A. Incidence and mortality of various cancers in iran and compare to other countries: a review article. Iran J Public Health, 2018;47(3):309–316.
Saberi M, et al. An In silico method to identify key proteins involved in the development of gastric cancer. Res Med. 2017;41(3):199–209.
Yousefi H, et al. Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene. 2019:1–22.
Dong P, et al. Long non-coding RNA NEAT1: a novel target for diagnosis and therapy in human tumors. Front Genet. 2018;9:471.
Powers J, et al. Short hairpin RNAs artifactually impair cell growth and suppress clustered microRNA expression. bioRxiv. 2018:372920.
Hu X, et al. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumor Biol. 2016;37(3):3497–504.
Yu X, Li Z. The role of microRNAs expression in laryngeal cancer. Oncotarget. 2015;6(27):23297.
Ding N, et al. NEAT1 regulates cell proliferation and apoptosis of ovarian cancer by miR-34a-5p/BCL2. Onco Targets Ther. 2017;10:4905–15.
Zhong F, et al. LncRNA NEAT1 promotes colorectal cancer cell proliferation and migration via regulating glial cell-derived neurotrophic factor by sponging miR-196a-5p. Acta Biochim Biophys Sin. 2018;50(12):1190–9.
Li J-H, et al. Long non-coding RNA NEAT1 promotes malignant progression of thyroid carcinoma by regulating miRNA-214. Int J Oncol. 2017;50(2):708–16.
Chakravarty D, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5(1):1–16.
He C, et al. Aberrant NEAT 1 expression is associated with clinical outcome in high grade glioma patients. APMIS. 2016;124(3):169–74.
Fu J-W, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142(7):1571–9.
Zhang J, et al. Silence of long noncoding RNA NEAT1 inhibits malignant biological behaviors and chemotherapy resistance in gastric cancer. Pathol Oncol Res. 2018;24(1):109–13.
Ma Y, et al. Enhanced expression of long non-coding RNA NEAT1 is associated with the progression of gastric adenocarcinomas. World J Surg Oncol. 2016;14(1):1–6.
Zhan T, et al. CRISPR/Cas9 for cancer research and therapy. in Seminars in cancer biology. Elsevier; 2019.
Liu C, et al. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17–26.
Yoshiba T, et al. CRISPR/Cas9-mediated cervical cancer treatment targeting human papillomavirus E6. Oncol Lett. 2019;17(2):2197–206.
Mehravar M, et al. Efficient Production of Biallelic RAG1 Knockout mouse embryonic stem cell using CRISPR/Cas9. Iran J Biotechnol. 2019;17(1):e2205.
Zhu S, et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR–Cas9 library. Nat Biotechnol. 2016;34(12):1279–86.
Skripova V, et al. CRISPR/Cas9 technique for identification of genes regulating oxaliplatin resistance of pancreatic cancer cell line. BioNanoScience. 2017;7(1):97–100.
Vitiello M, Tuccoli A, Poliseno L. Long non-coding RNAs in cancer: implications for personalized therapy. Cell Oncol. 2015;38(1):17–28.
Yu X, et al. NEAT 1: A novel cancer-related long non-coding RNA. Cell Prolif. 2017;50(2):e12329.
Luo Y, et al. Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/β-catenin signaling pathway. Cancer Lett. 2019;440:11–22.
Shin VY, et al. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019;10(4):1–10.
Qi L, et al. lncRNA NEAT1 competes against let-7a to contribute to non-small cell lung cancer proliferation and metastasis. Biomed Pharmacother. 2018;103:1507–15.
Li Y, et al. LncRNA NEAT1 regulates cell viability and invasion in esophageal squamous cell carcinoma through the miR-129/CTBP2 axis. Dis Markers. 2017;2017:1–12.
Cossu AM, et al. Long non-coding RNAs as important biomarkers in laryngeal cancer and other head and neck tumours. Int J Mol Sci. 2019;20(14):3444.
Zhang Y-Q, et al. CRISPR/Cas9-mediated knockout of the PDEF gene inhibits migration and invasion of human gastric cancer AGS cells. Biomed Pharmacother. 2019;111:76–85.
Li Y, et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6(29):27641.
Hu Y, et al. Knockdown of the oncogene lncRNA NEAT1 restores the availability of miR-34c and improves the sensitivity to cisplatin in osteosarcoma. Biosci Rep. 2018;38(3):1–10.
Acknowledgements
The results presented in this paper were part of the Ph.D student thesis and were financially supported by personal student cost.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Haghighi, N., Doosti, A. & Kiani, J. Evaluation of CRISPR/Cas9 System Effects on Knocking Out NEAT1 Gene in AGS Gastric Cancer Cell Line with Therapeutic Perspective. J Gastrointest Canc 53, 623–631 (2022). https://doi.org/10.1007/s12029-021-00669-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-021-00669-z