Abstract
Background
The 1245C>G (rs1052133) polymorphism of human 8-oxoguanine DNA glycosylase 1 (hOGG1) gene has been indicated to be correlated with colorectal (CRC) susceptibility, but studies have yielded conflicting results. Thus, the present meta-analysis was performed to derive a more precise estimation between hOGG1 1245C>G polymorphism and CRC risk.
Methods
Data were collected from several electronic databases such as PubMed, EMBASE, and Google Scholar databases, with the last search up to September 01, 2020. Pooled odds ratios (ORs) with corresponding 95% confidence intervals (CIs) were used to assess the strength of the association.
Results
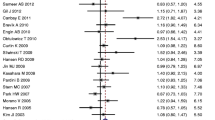
A total of 24 case-control studies with 7010 CRC cases and 10,674 controls were selected. Pooled data showed that the hOGG1 1245C>G polymorphism was significantly associated with CRC risk under three genetic models, i.e., homozygote (GG vs. CC: OR = 1.229, 95% CI 1.031–1.465, p = 0.022); heterozygote (GC vs. CC: OR = 1.142, 95% CI 1.008–1.294, p = 0.037); and dominant (GG+GC vs. CC: OR = 1.162, 95% CI 1.034–1.304, p = 0.011). When stratified analysis by ethnicity, a significant association of the hOGG1 1245C>G polymorphism with risk of CRC was found in the Caucasians, but not in Asians. Moreover, there were significant associations between hOGG1 1245C>G polymorphism and CRC by PCR-RFLP and hospital-based subgroups.
Conclusions
Inconsistent with the previous meta-analysis, these meta-analysis results revealed that the hOGG1 1245C>G polymorphism might be associated with an increased risk of CRC, especially in Caucasians.



Similar content being viewed by others
References
Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. Thieme Medical Publishers. 2009;22:191–7.
Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14:89–103.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–91.
Bishehsari F, Mahdavinia M, Vacca M, Malekzadeh R, Mariani-Costantini R. Epidemiological transition of colorectal cancer in developing countries: environmental factors, molecular pathways, and opportunities for prevention. World J Gastroenterol. 2014;20:6055–72.
Valle L, de Voer RM, Goldberg Y, Sjursen W, Försti A, Ruiz-Ponte C, et al. Update on genetic predisposition to colorectal cancer and polyposis. Mol Asp Med. 2019;69:10–26.
Valle L, Vilar E, Tavtigian SV, Stoffel EM. Genetic predisposition to colorectal cancer: syndromes, genes, classification of genetic variants and implications for precision medicine. J Pathol. 2019;247:574–88.
Kastrinos F, Syngal S. Inherited colorectal cancer syndromes. Cancer J. 2011;17:405–15.
Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–58.
Ba X, Boldogh L. 8-Oxoguanine DNA glycosylase 1: beyond repair of the oxidatively modified base lesions. Redox Biol. 2018;14:669–78.
Wang R, Hao W, Pan L, Boldogh I, Ba X. The roles of base excision repair enzyme OGG1 in gene expression. Cell Mol Life Sci. Birkhauser Verlag AG. 2018:3741–50.
Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol Cell Biol. 2006;26:1654–65.
Ali K, Mahjabeen I, Sabir M, Mehmood H, Kayani MA. OGG1 mutations and risk of female breast cancer: meta-analysis and experimental data. Dis Markers. 2015;2015:1–16.
Alanazi M, Pathan AAK, Ajaj SA, Khan W, Shaik JP, Al Tassan NA, et al. DNA repair genes XRCC1, XRCC3, XPD, AND OGG1 polymorphisms among the central region population of Saudi Arabia. Biol Res. Sociedad de Biología de Chile. 2013;46:161–7.
Shinmura K, Yokota J. The OGG1 gene encodes a repair enzyme for oxidatively damaged DNA and is involved in human carcinogenesis. Antioxid Redox Signal. Mary Ann Liebert Inc. 2001;3:597–609.
Sebera J, Hattori Y, Sato D, Reha D, Nencka R, Kohno T, et al. The mechanism of the glycosylase reaction with hOGG1 base-excision repair enzyme: concerted effect of Lys249 and Asp268 during excision of 8-oxoguanine. Nucleic Acids Res. Oxford University Press. 2017;45:5231–42.
Zou H, Li Q, Xia W, Liu Y, Wei X, Wang D. Association between the OGG1 Ser326Cys polymorphism and cancer risk: evidence from 152 case-control studies. J Cancer. Ivyspring International Publisher. 2016;7:1273–80.
Zhang Y, He B-S, Pan Y-Q, Xu Y-Q, Wang S-K. Association of OGG1 Ser326Cys polymorphism with colorectal cancer risk: a meta-analysis. Int J Color Dis. 2011;26:1525–30.
Kim J-I, Park Y-J, Kim K-H, Kim J-I, Song B-J, Lee M-S, et al. hOGG1 Ser326Cys polymorphism modifies the significance of the environmental risk factor for colon cancer. World J Gastroenterol. 2003;9:956–60.
Hansen R, Sæbø M, Skjelbred CF, Nexø BA, Hagen PC, Bock G, et al. GPX Pro198Leu and OGG1 Ser326Cys polymorphisms and risk of development of colorectal adenomas and colorectal cancer. Cancer Lett. 2005;229:85–91.
Hansen RD, Krath BN, Frederiksen K, Tjønneland A, Overvad K, Roswall N, et al. GPX1 Pro198Leu polymorphism, erythrocyte GPX activity, interaction with alcohol consumption and smoking, and risk of colorectal cancer. Mutat Res Fundam Mol Mech Mutagen. 2009;664:13–9.
Engin AB, Karahalil B, Engin A, Karakaya AE. Oxidative stress, Helicobacter pylori, and OGG1 Ser326Cys, XPC Lys939Gln, and XPD Lys751Gln polymorphisms in a Turkish population with colorectal carcinoma. Genet Test Mol Biomarkers. 2010;14:559–64.
Obtulowicz T, Swoboda M, Speina E, Gackowski D, Rozalski R, Siomek A, et al. Oxidative stress and 8-oxoguanine repair are enhanced in colon adenoma and carcinoma patients. Mutagenesis. 2010;25:463–71.
Brevik A, Joshi AD, Corral R, Onland-Moret NC, Siegmund KD, Le Marchand L, et al. Polymorphisms in base excision repair genes as colorectal cancer risk factors and modifiers of the effect of diets high in red meat. Cancer Epidemiol Biomark Prev. 2010;19:3167–73.
Canbay E, Cakmakoglu B, Zeybek U, Sozen S, Cacina C, Gulluoglu M, et al. Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population. Curr Med Res Opin. 2011;27:1295–302.
Gil J, Ramsey D, Stembalska A, Karpinski P, Pesz KA, Laczmanska I, et al. The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual’s susceptibility to sporadic colorectal cancer. Mol Biol Rep. 2012;39:527–34.
Sameer AS, Nissar S, Abdullah S, Chowdri NA, Siddiqi MA. DNA repair gene 8-oxoguanine DNA glycosylase Ser326Cys polymorphism and colorectal cancer risk in a Kashmiri population. DNA Cell Biol. 2012;31:541–6.
Yan J, Mingwei Y, Zhaoya Y, Yuhua K, Jinbo D, Bin L. The relationship between hOGG1 and XPD gene polymorphism and genetic susceptibility of digestive system tumors. Chin J Clin Oncol. 2012;39:1358–62.
Przybylowska K, Kabzinski J, Sygut A, Dziki L, Dziki A, Majsterek I. An association selected polymorphisms of XRCC1, OGG1 and MUTYH gene and the level of efficiency oxidative DNA damage repair with a risk of colorectal cancer. Mutat Res Fundam Mol Mech Mutagen. 2013;745–746:6–15.
Zhang S-H, Wang L-A, Li Z, Peng Y, Cun Y-P, Dai N, et al. APE1 polymorphisms are associated with colorectal cancer susceptibility in Chinese Hans. World J Gastroenterol. 2014;20:8700.
Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, Blanco I, et al. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101–8.
Lai C-Y, Hsieh L-L, Tang R, Santella RM, Chang-Chieh CR, Yeh C-C. Association between polymorphisms of APE1 and OGG1 and risk of colorectal cancer in Taiwan. World J Gastroenterol. 2016;22:3372–80.
Zhang Q, Zheng X, Li X, Sun D, Xue P, Zhang G, et al. The polymorphisms of miRNA-binding site in MLH3 and ERCC1 were linked to the risk of colorectal cancer in a case-control study. Cancer Med. 2018;7:1264–74.
Kabzinski J, Walczak A, Dziki A, Mik M, Majsterek I. Impact of the Ser326Cys polymorphism of the OGG1 gene on the level of oxidative DNA damage in patients with colorectal cancer. Pol J Surg. 2018;90:13–5.
Park H-W, Kim I-J, Kang HC, Jang S-G, Ahn S-A, Lee JS, et al. The hOGG1 Ser326Cys polymorphism is not associated with colorectal cancer risk. J Epidemiol. 2007;17:156–60.
Stern MC, Conti DV, Siegmund KD, Corral R, Yuan J-M, Koh W-P, et al. DNA repair single-nucleotide polymorphisms in colorectal cancer and their role as modifiers of the effect of cigarette smoking and alcohol in the Singapore Chinese Health Study. Cancer Epidemiol Biomark Prev. 2007;16:2363–72.
Kasahara M, Osawa K, Yoshida K, Miyaishi A, Osawa Y, Inoue N, et al. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. J Exp Clin Cancer Res. 2008;27:49.
Pardini B, Naccarati A, Novotny J, Smerhovsky Z, Vodickova L, Polakova V, et al. DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res Fundam Mol Mech Mutagen. 2008;638:146–53.
Curtin K, Samowitz WS, Wolff RK, Ulrich CM, Caan BJ, Potter JD, et al. Assessing tumor mutations to gain insight into base excision repair sequence polymorphisms and smoking in colon cancer. Cancer Epidemiol Biomark Prev. 2009;18:3384–8.
Ming-juan J, Bing L, Shuang-shuang Z, Yong-jing Z, Mei X, Xin-yuan M, et al. Application of multifactor dimensionality reduction on the interactions between gene-gene, geneenvironment and the risk sporadic colorectal cancer in Chinese population. Chin J Epidemiol. 2008;29:535–9.
Aizhong W, Wenming C, Xianglei H, Hangruo J, Xiaxiang J, Guanshan Z, et al. A hOGG1 gene polymorphism and genetic susceptibility to colorectal cancer and hepatocellular carcinoma. Chin J Gastroenterol Hepatol. 2008;17:854–7.
Sliwinski T, Krupa R, Wisniewska-Jarosinska M, Pawlowska E, Lech J, Chojnacki J, et al. Common polymorphisms in the XPD and hOGG1 genes are not associated with the risk of colorectal cancer in a Polish population. Tohoku J Exp Med. 2009;218:185–91.
Sun X, Yang H, Lin Y, Zhao J, Bao Y, Liu X, et al. Genetic association between hOGG1 C8069G polymorphism and colorectal cancer risk. Int J Clin Exp Med. 2015;8:21145–51.
Wang Y, Gao X, Wei F, Zhang X, Yu J, Zhao H, et al. The hOGG1 Ser326Cys polymorphism contributes to digestive system cancer susceptibility: evidence from 48 case–control studies. Tumor Biol. 2015;36:1029–38.
Sobhan MR, Mahdinezhad-Yazdi M, Dastgheib SA, Jafari M, Raee-Ezzabadi A, Neamatzadeh H. Association of ESRα XbaI A>G, ESRα PvuII T>C and ESRβ AlwNI T>C polymorphisms with the risk of developing adolescent idiopathic scoliosis: a systematic review and genetic meta-analysis. Rev Bras Ortop. 2020;55:8–16.
Ferdosian F, Jarahzadeh MH, Bahrami R, Nafei Z, Jafari M, Raee-Ezzabadi A, et al. Association of IL-6 -174G>C polymorphism with susceptibility to childhood sepsis: a systematic review and meta-analysis. Fetal Pediatr Pathol. 2020:1–15 Online ahead of print.
Bahrami R, Dastgheib SA, Niktabar SM, Amooee A, Lookzadeh MH, Mirjalili SR, et al. Association of BMP4 rs17563 polymorphism with nonsyndromic cleft lip with or without cleft palate risk: literature review and comprehensive meta-analysis. Fetal Pediatr Pathol. 2020:1–15 Online ahead of print.
Acknowledgments
I would like to express my sincere gratitude to Professor Mohammad Hasan Sheikhha for his motivation, knowledge, and support during the course of this research.
Author information
Authors and Affiliations
Contributions
Y.G. and F.A. are responsible as the guarantor of integrity of the entire study, study design and concepts, definition of intellectual content, and literature research. M.H.A. and S.A.D. are responsible for the clinical studies, experimental studies, data acquisition, and manuscript preparation. S.H.S and J.J.N. are responsible for the data analysis, statistical analysis, and manuscript review. H.N and Y.G are responsible for the manuscript editing. All authors have read and agreed with the final version of this manuscript.
Corresponding author
Ethics declarations
An ethical approval or patient consent was not needed because this is a meta-analysis in which all data were extracted from published literature.
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ghelmani, Y., Asadian, F., Antikchi, M.H. et al. Association Between the hOGG1 1245C>G (rs1052133) Polymorphism and Susceptibility to Colorectal Cancer: a Meta-analysis Based on 7010 Cases and 10,674 Controls. J Gastrointest Canc 52, 389–398 (2021). https://doi.org/10.1007/s12029-020-00532-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-020-00532-7