Abstract
Background
Allergic rhinitis (AR) is a common allergic disease with increasing prevalence globally. However, the molecular mechanism underlying AR pathogenesis remains largely undefined.
Methods
Peripheral blood and nasal mucosa samples obtained from patients with AR (n = 22), and ovalbumin-induced AR mouse model (n = 8 per group) were prepared for subsequent detection. qRT-PCR and western blot were used to detect the expression of LINC00240, miR-155-5p, PU.1 and other key molecules. ELISA assay and flow cytometry were employed to evaluate the secretion of IL-9 and T-helper 9 (Th9) cell ratio, respectively. Bioinformatics analysis, RNA immunoprecipitation (RIP), chromatin immunoprecipitation (ChIP) and luciferase reporter assays were employed to further elucidate the regulatory network of LINC00240/miR-155-5p/DNMT1. The methylation of PU.1 promoter was assessed by methylation-specific PCR (MSP). This signaling axis was further validated in the mouse model of AR.
Results
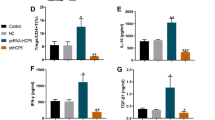
LINC00240 was downregulated, while miR-155-5p and PU.1 were upregulated in the peripheral blood and nasal mucosa of AR patients, as well as in AR mice. This was accompanied with the increased ratio of Th9 cells and elevated IL-9 secretion. Mechanistically, LINC00240 served as a miR-155-5p sponge, and DNMT1 was a target of miR-155-5p. In addition, DNMT1 mediated the methylation of PU.1 promoter. In vivo studies verified that LINC00240 mitigated AR progression, possibly via miR-155-5p/DNMT1/PU.1-dependent Th9 differentiation.
Conclusion
The involvement of LINC00240 in AR pathogenesis is closely associated with Th9 differentiation through modulating DNMT1-dependent methylation of PU.1 by sponging miR-155-5p.
Highlights
LINC00240 was downregulated, while miR-155-5p and PU.1 were upregulated in the serum and nasal mucosa of AR patients and mice.
The percentage of Th9 cells and IL-9 secretion were elevated in AR patients and mice.
LINC00240 served as a miR-155-5p sponge, and DNMT1 was a target of miR-155-5p.
DNMT1 mediated the methylation of PU.1 promoter.
LINC00240/miR-155-5p/DNMT1/PU.1 axis is involved in AR pathogenesis via modulating Th9 differentiation.










Similar content being viewed by others
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Abbreviations
- 5-Aza:
-
5-Azacytidine
- AD:
-
Atopic dermatitis
- AR:
-
Allergic rhinitis
- ChIP:
-
Chromatin immunoprecipitation
- DNMT:
-
DNA methyltransferase
- ELISA:
-
Enzyme-linked immunosorbent assay
- FISH:
-
Fluorescence in situ hybridization
- GC:
-
Gastric cancer
- HCC:
-
Hepatocellular carcinoma
- H&E:
-
Hematoxylin & Eosin
- IgE:
-
Immunoglobulin E
- IL-4:
-
Interleukin-4
- ILC2s:
-
Group 2 innate lymphoid cells
- lncRNA:
-
Long non-coding RNA
- MSP:
-
Methylation-specific PCR
- MUT:
-
Mutated
- OVA:
-
Ovalbumin
- NPC:
-
Nasopharyngeal carcinoma
- NSCLC:
-
Non-small cell lung cancer
- RIP:
-
RNA immunoprecipitation
- SPF:
-
Specific-pathogen-free
- TGF-β:
-
Transforming growth factor-β
- Th9:
-
T-helper 9
- WT:
-
Wild type
References
Hoyte FCL, Nelson HS. Recent advances in allergic rhinitis. F1000Res. 2018;7. https://doi.org/10.12688/f1000research.15367.1.
Meng Y, Wang C, Zhang L. Recent developments and highlights in allergic rhinitis. Allergy. 2019;74(12):2320–8. https://doi.org/10.1111/all.14067.
Ciprandi G, De Amici M, Murdaca G, Fenoglio D, Ricciardolo F, Marseglia G, et al. Serum interleukin-17 levels are related to clinical severity in allergic rhinitis. Allergy. 2009;64(9):1375–8. https://doi.org/10.1111/j.1398-9995.2009.02010.x.
Pilette C, Jacobson MR, Ratajczak C, Detry B, Banfield G, VanSnick J, et al. Aberrant dendritic cell function conditions Th2-cell polarization in allergic rhinitis. Allergy. 2013;68(3):312–21. https://doi.org/10.1111/all.12090.
Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, et al. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9(12):1341–6. https://doi.org/10.1038/ni.1659.
Angkasekwinai P. Th9 cells in allergic Disease. Curr Allergy Asthma Rep. 2019;19(5):29. https://doi.org/10.1007/s11882-019-0860-8.
Kaplan MH, Hufford MM, Olson MR. The development and in vivo function of T helper 9 cells. Nat Rev Immunol. 2015;15(5):295–307. https://doi.org/10.1038/nri3824.
Gerlach K, Hwang Y, Nikolaev A, Atreya R, Dornhoff H, Steiner S, et al. TH9 cells that express the transcription factor PU.1 drive T cell-mediated Colitis via IL-9 receptor signaling in intestinal epithelial cells. Nat Immunol. 2014;15(7):676–86. https://doi.org/10.1038/ni.2920.
Wang XQ, Hu GH, Kang HY, Shen Y, Ke X, Hong SL. High frequency of T helper type 9 cells in Chinese patients with allergic rhinitis. Asian Pac J Allergy Immunol. 2015;33(4):301–7. https://doi.org/10.12932/AP0609.33.4.2015.
Jiang X, Zhang X, Liu J, Liu J, Zhu X, Yang C. Involvement of T-Helper 9 activation in a mouse model of allergic Rhinitis. Med Sci Monit. 2018;24:4704–10. https://doi.org/10.12659/MSM.908302.
Li L, Deng J, Huang T, Liu K, Jiang X, Chen X, et al. IRF4 transcriptionally activate HOTAIRM1, which in turn regulates IRF4 expression, thereby affecting Th9 cell differentiation and involved in allergic rhinitis. Gene. 2022;813:146118. https://doi.org/10.1016/j.gene.2021.146118.
DiStefano JK. The emerging role of long noncoding RNAs in Human Disease. Methods Mol Biol. 2018;1706:91–110. https://doi.org/10.1007/978-1-4939-7471-9_6.
Wei X, Xu M, Wang C, Fang S, Zhang Y, Wang W. Genome-wide analysis of long noncoding RNA expression profile in nasal mucosa with allergic rhinitis. BMC Med Genomics. 2021;14(1):100. https://doi.org/10.1186/s12920-021-00949-4.
Bu WJ, Fang Z, Li WL, Wang X, Dong MJ, Tao QY, et al. LINC00240 sponges miR-4465 to promote proliferation, migration, and invasion of hepatocellular carcinoma cells via HGF/c-MET signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(20):10452–61. https://doi.org/10.26355/eurrev_202010_23397.
Zhang Y, Li X, Zhang J, Liang H. Natural killer T cell cytotoxic activity in Cervical cancer is facilitated by the LINC00240/microRNA-124-3p/STAT3/MICA axis. Cancer Lett. 2020;474:63–73. https://doi.org/10.1016/j.canlet.2019.12.038.
Ku GW, Kang Y, Yu SL, Park J, Park S, Jeong IB, et al. LncRNA LINC00240 suppresses invasion and migration in non-small cell Lung cancer by sponging miR-7-5p. BMC Cancer. 2021;21(1):44. https://doi.org/10.1186/s12885-020-07755-8.
Wang G, Zhang Z, Xia C. Long non-coding RNA LINC00240 promotes gastric cancer progression via modulating miR-338-5p/METTL3 axis. Bioengineered. 2021;12(2):9678–91. https://doi.org/10.1080/21655979.2021.1983276.
Chen X, Wu G, Qing J, Li C, Chen X, Shen J. LINC00240 knockdown inhibits nasopharyngeal carcinoma progress by targeting miR-26a-5p. J Clin Lab Anal. 2022;36(5):e24424. https://doi.org/10.1002/jcla.24424.
Cheng S, Tang Q, Xie S, Wen S, Zhang H, Xie Z, et al. The role of noncoding RNA in Airway Allergic Diseases through Regulation of T Cell subsets. Mediators Inflamm. 2022;2022:6125698. https://doi.org/10.1155/2022/6125698.
Wu XH, Zhao SJ, Huang WQ, Huang LH, Luo XY, Long SL. Long non-coding RNA MALAT1 promotes Th2 differentiation by regulating microRNA-135b-5p/GATA-3 axis in children with allergic rhinitis. Kaohsiung J Med Sci. 2022;38(10):971–80. https://doi.org/10.1002/kjm2.12587.
Yang C, Shangguan C, Cai C, Xu J, Qian X. LncRNA HCP5 participates in the Tregs functions in allergic rhinitis and drives Airway Mucosal Inflammatory response in the nasal epithelial cells. Inflammation. 2022;45(3):1281–97. https://doi.org/10.1007/s10753-022-01620-5.
Suojalehto H, Toskala E, Kilpelainen M, Majuri ML, Mitts C, Lindstrom I, et al. MicroRNA profiles in nasal mucosa of patients with allergic and nonallergic rhinitis and Asthma. Int Forum Allergy Rhinol. 2013;3(8):612–20. https://doi.org/10.1002/alr.21179.
Malmhall C, Alawieh S, Lu Y, Sjostrand M, Bossios A, Eldh M, et al. MicroRNA-155 is essential for T(H)2-mediated allergen-induced eosinophilic inflammation in the lung. J Allergy Clin Immunol. 2014;133(5):1429–38. https://doi.org/10.1016/j.jaci.2013.11.008.
Zhu YQ, Liao B, Liu YH, Wang Z, Zhu XH, Chen XB, et al. MicroRNA-155 plays critical effects on Th2 factors expression and allergic inflammatory response in type-2 innate lymphoid cells in allergic rhinitis. Eur Rev Med Pharmacol Sci. 2019;23(10):4097–109. https://doi.org/10.26355/eurrev_201905_17911.
Hammad NM, Nabil F, Elbehedy EM, Sedeek R, Gouda MI, Arafa MA, et al. Role of MicroRNA-155 as a potential biomarker for allergic Rhinitis in Children. Can Respir J. 2021;2021:5554461. https://doi.org/10.1155/2021/5554461.
Zhu Y, Ye F, Fu Y, Zhu X, Wang Z, Wu S, et al. MicroRNA-155-5p regulates the Th1/Th2 cytokines expression and the apoptosis of group 2 innate lymphoid cells via targeting TP53INP1 in allergic rhinitis. Int Immunopharmacol. 2021;101(Pt B):108317. https://doi.org/10.1016/j.intimp.2021.108317.
Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010;11(6):527–34. https://doi.org/10.1038/ni.1867.
Scadding GK, Kariyawasam HH, Scadding G, Mirakian R, Buckley RJ, Dixon T, et al. BSACI guideline for the diagnosis and management of allergic and non-allergic rhinitis (revised Edition 2017; First Edition 2007). Clin Exp Allergy. 2017;47(7):856–89. https://doi.org/10.1111/cea.12953.
Cordero P, Campion J, Milagro FI, Goyenechea E, Steemburgo T, Javierre BM, et al. Leptin and TNF-alpha promoter methylation levels measured by MSP could predict the response to a low-calorie diet. J Physiol Biochem. 2011;67(3):463–70. https://doi.org/10.1007/s13105-011-0084-4.
Agache I, Akdis CA. Precision medicine and phenotypes, endotypes, genotypes, regiotypes, and theratypes of allergic Diseases. J Clin Invest. 2019;129(4):1493–503. https://doi.org/10.1172/JCI124611.
Eifan AO, Durham SR. Pathogenesis of rhinitis. Clin Exp Allergy. 2016;46(9):1139–51. https://doi.org/10.1111/cea.12780.
Georas SN, Guo J, De Fanis U, Casolaro V. T-helper cell type-2 regulation in allergic Disease. Eur Respir J. 2005;26(6):1119–37. https://doi.org/10.1183/09031936.05.00006005.
Hoppenot D, Malakauskas K, Lavinskiene S, Bajoriuniene I, Kalinauskaite V, Sakalauskas R. Peripheral blood Th9 cells and eosinophil apoptosis in Asthma patients. Med (Kaunas). 2015;51(1):10–7. https://doi.org/10.1016/j.medici.2015.01.001.
Shin JH, Kim DH, Kim BY, Kim SW, Hwang SH, Lee J, et al. Anti-interleukin-9 antibody increases the Effect of Allergen-Specific Immunotherapy in Murine allergic rhinitis. Allergy Asthma Immunol Res. 2017;9(3):237–46. https://doi.org/10.4168/aair.2017.9.3.237.
Galli SJ, Tsai M. IgE and mast cells in allergic Disease. Nat Med. 2012;18(5):693–704. https://doi.org/10.1038/nm.2755.
Ma Z, Teng Y, Liu X, Li J, Mo J, Sha M, et al. Identification and functional profiling of differentially expressed long non-coding RNAs in nasal mucosa with allergic Rhinitis. Tohoku J Exp Med. 2017;242(2):143–50. https://doi.org/10.1620/tjem.242.143.
Ma Y, Shi L, Zheng C. Microarray analysis of lncRNA and mRNA expression profiles in mice with allergic rhinitis. Int J Pediatr Otorhinolaryngol. 2018;104:58–65. https://doi.org/10.1016/j.ijporl.2017.10.046.
Ma Y, Shi L, Zhao K, Zheng C. lncRNA FR215775 regulates Th2 differentiation in Murine allergic rhinitis. J Immunol Res. 2022;2022:7783481. https://doi.org/10.1155/2022/7783481.
Ma Z, Lian H, Lin X, Li Y. LncRNA MIAT promotes allergic inflammation and symptoms by targeting MiR-10b-5p in allergic Rhinitis mice. Am J Rhinol Allergy. 2021;35(6):781–9. https://doi.org/10.1177/1945892421998143.
Wu HY, Liu K, Zhang JL. LINC00240/miR-155 axis regulates function of trophoblasts and M2 macrophage polarization via modulating oxidative stress-induced pyroptosis in preeclampsia. Mol Med. 2022;28(1):119. https://doi.org/10.1186/s10020-022-00531-3.
Srivastava M, Kaplan MH. Transcription factors in the development and pro-allergic function of mast cells. Front Allergy. 2021;2:679121. https://doi.org/10.3389/falgy.2021.679121.
Hamza AM, Omar SS, Abo El-Wafa RA, Elatrash MJ. Expression levels of transcription factor PU.1 and interleukin-9 in atopic dermatitis and their relation to Disease severity and eruption types. Int J Dermatol. 2017;56(5):534–9. https://doi.org/10.1111/ijd.13579.
Ai S, Lin Y, Zheng J, Zhuang X. Xingbi Gel ameliorates allergic Rhinitis by regulating IFN-gamma gene promoter methylation in CD4 + T cells via the ERK-DNMT pathway. Front Surg. 2020;7:619053. https://doi.org/10.3389/fsurg.2020.619053.
Cui X, Guo Y, Wang Q, Li X. MiR-199-3p-Dnmt3a-STAT3 signalling pathway in ovalbumin-induced allergic rhinitis. Exp Physiol. 2019;104(8):1286–95. https://doi.org/10.1113/EP087751.
Acknowledgements
We would like to give our sincere gratitude to the reviewers for their constructive comments.
Funding
This study was funded by the Applied Research training program of Science and Technology Department of Jiangxi province (No.20212BAG70031), the National Natural Science Foundation of China (No. 82160211) and the Jiangxi Provincial Natural Science Foundation (No. 20202ACBL206013).
Author information
Authors and Affiliations
Contributions
Conception and design of study: C. Y. Acquisition of data: J. L.; K. L.; Y. Q. Analysis and interpretation of data: X. J.; J. D.; W. W. Drafting the manuscript: J. L. Revising the manuscript critically for important intellectual content: C. Y. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study has obtained approval of the Ethics Committee of The Second Affiliated Hospital of Nanchang University and the patient’s written informed consent.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, J., Jiang, X., Liu, K. et al. Role of LINC00240 on T-helper 9 differentiation in allergic rhinitis through influencing DNMT1-dependent methylation of PU.1. Immunol Res 72, 197–211 (2024). https://doi.org/10.1007/s12026-023-09435-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-023-09435-8