Abstract
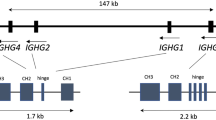
The IGHG (ImmunoGlobulin constant Heavy G chain) genes are situated close to the IGHE gene on chromosome 14q32, 5′μ, δ, γ3, γ1, α1, γ2, γ4, ε, α2, 3′, in linkage disequilibrium. The polymorphism of γ3, γ1 and γ2 genes, is investigated as alternative allotypes. They are inherited in a Mendelian fashion and are expressed randomly in allelic exclusion. The alternative and functionally different γ3, γ1 and γ2 gene variants, are found in four IGHG haplotypes, coding 4 B-cell variants: IGHG*bfn (=B1-cells), IGHG*bf-n (=B2-cells), IGHG*gan (=B3-cells) and IGHG*ga-n (=B4-cells). The dominance of the IGHG2*n allele from the IGHG*bfn haplotype (=B1-cells) has been shown in repeated investigations, namely in patients with asthma and allergy with increased serum levels of IgE > 600 ku/l and more often so in those with IgE > 1,000 ku/l or IgG4>1 g/l, in childhood asthma patients with mean level of IgE = 1,762 ku/l and in allergen exposed individuals developing laboratory animal allergy. In children with non-atopy and mean IgE level = 9.5 ku/l there is instead a dominance of the alternative allotypes from the IGHG*ga-n (=B4-cells) with IGHG2*-n alleles. In a case–control study allergic children with a family history of allergy, clinically manifest allergy and/or positive SPT, the IGHG*bfn haplotype (=B1-cells) with the IGHG2*n allele dominates, with increased risk of atopy and the IGHG*bf-n haplotype (=B2-cells) with the IGHG2*-n allele is infrequent with low risk, probably protective against atopy. The phenotypic expressions of the IGHG*bfn haplotype (=B1 cells) and IGHG*bfn/*bfn diplotypes (B1/B1-cells) are increased IgG2*n allotype together with increased IgE serum levels and IgE sensitisation in agreement with atopy. The alternative IGHG*ga-n/*ga-n diplotype (B4/B4-cells) express low IgG1*a- and IgG2*-n allotypes, together with low IgE and non-IgE sensitisation, in agreement with non-atopy. Together these studies have given us a greater understanding of the involvement of IGHG genes, IGHG coded B-cells and immunochemical and functional variants of IgG molecules describing different forms of asthma and allergy, which will improve diagnoses and treatment.
Similar content being viewed by others
References
Cookson WO. Genetic and genomics of asthama and allergic diseases. Immunol Rev 2002;190(1):195–206
Grubb R. Human immunoglobulin allotypes and Mendelian polymorphism of the human immunoglobulin genes. In: Oss CJ, Regenmortel MHV, editors. Immunochemistry. New York: Dekker; 1994. p 47–68
Oxelius V-A. Correlation between atopy and Gm allotypes. Int Arch Allergy Appl Immunol 1990;91:54–7
Oxelius V-A, Hultquist C, Husby S. Gm allotypes as indicators of non-atopic and atopic bronchial asthma. Int Arch Allergy Appl Immunol 1993;101:66–71
Oxelius V-A. Gm allotype genes and gene dosage affecting both IgG subclass and IgE levels in atopic patients. Int Arch Allergy Appl Immunol 1990;91:58–61
Oxelius V-A, Carlsson A-M. Quantitation of Gm allotypes. Scand J Immunol 1993;37:143–8
Oxelius V-A. Preparation of IgG subclass allotypes from polyclonal IgG. Scand J Immunol 1999;49:395–8
Oxelius V-A, Eibl MM. Different Gm allotype levels in human intravenous immunglobulin (IVIG) preparations, survival of foreign Gm allotypes in immunodeficient patients. Clin Exp Immunol 1996;106:203–7
Oxelius V-A. Genetic B cell variation based on immunoglobulin heavy G chain (Gm) genes. Scand J Immunol 1999;49:345–6
Oxelius V-A. Serum IgG and IgG subclass contents in different Gm phenotypes. Scand J Immunol 1993;37:149–53
Oxelius V-A, Aurivillius M, Carlsson A-M, Musil K. Serum Gm allotype development during childhood. Scand J Immunol 1999;50:440–6
Oxelius V-A. Lack of the G2m(n) allotype in IgG subclass deficiency, in IgG2 deficiency together with lack of G1m(a) and G3m(g) and IgG3 deficiency together with lack of G1m(f) and G3m(b). Scand J Immunol 1990;31:243–7
Cookson WO. The alliance of genes and environment in asthma and allergy. Nature 1999;402 Suppl:B5–11
Spaich D, Ostertag M. Untersuchungen uber allergische Erkrankungen bei Zwillingen. Z Mensch Vererb-Konstitutionsl 1936;19:731–51
Schultz-Larsen F, Holm NV, Henigsen K. Atopic dermatitis: a genetic-epidemiologic study in a population-based twin sample. J Am Acad Dermatol 1986;15:487–94
Oxelius V-A, Sjöstedt L, Willers S, Löw B. Development of allergy to laboratory animals is associated with particular Gm and HLA genes. Int Arch Allergy Immunol 1996;110:73–8
Aurivillius M, Oymar K, Oxelius V-A. Immunoglobulin heavy G2 chain (IGHG2) gene restriction in the development of severe respiratory syncytial virus infection. Acta Paediatr 2005;94:414–8
Oxelius V-A, Carlsson A-M, Aurivillius M. Alternative G1m, G2m and G3m allotypes of IGHG genes correlate with atopic and non atopic pathways of immune regulation in children with bronchial asthma. Int Arch Allergy Immunol 1998;115:215–9
Oxelius V-A. Imbalanced switch of the IGHG (ImmunoGlobulin constant Heavy G chain) Gm(bfn) genes in atopic childhood asthma. Allergy 2000;55:1063–8
Walley AJ, Cookson WO. Investigation of an interleukin-4 promoter polymorphism for association with asthma and atopy. J Med Genetic 1996;33:689–92
Willis-Karp M, et al. Interleukin-13: central mediator of allergic asthma. Science 1998;282:2258–61
Pandey J, Fudenberg H, Virella G, et al. Association between immunoglobulin allotypes and immune responses to Haemophilus influenzae and meningococcus polysaccharides. Lancet 1979;I:190–2
Ambrosino D, Schiffman G, Gotschlich E, et al. Correlation between G2m(n) immunoglobulin allotype and human antibody response and susceptibility to polysaccharide encapsulated bacteria. J Clin Invest 1985;75:1935–42
Legrand L, Rivat-Perran L, Hutting C, Dausset J. HLA and Gm linked genes affecting the degradation of antigens (sheep red cells) endocytosed by macrophages. Hum Immunol 1982;4:1–13
Oxelius V-A, Hanson L-Å, Björkander J, Hammarström L, Sjöholm A. IgG3 deficiency: common in obstructive lung disease. Monogr Allergy 1986;20:106–15
ISAAC. Worldwide variation in prevalence of symptoms of asthma, allergic rhinoconjunctivitis and atopic eczema. ISAAC The international study of asrhma and allergies in childhood (ISAAC) steering committee. Lancet 1998;351:1225–32
Oxelius V-A, Bråbäck L, Ahlstedt S, Björkstén B. Immunoglobulin constant heavy G chain genes as risk factors in childhood allergies. Clin Exp Allergy 2006;36:1616–24
Jarvis D, Burney P. ABC of allergies. The epidemiology of allergic diseases. BMJ 1998;316:607–9
Holgate ST. The epidemic of allergy and asthma. Nature 1999;402:B2–4
Marsch DG, Meyers DA, Bias WB. The epidemiology and genetics of atopic allergy. New Engl J Med 1981;305:1551–9
Cookson WO, Sharp PA, Faux JA, Hopkin JM. Linkage between immunoglobulin E-responses underlying asthma and rhinitis and chromosome 11q. Lancet 1989;i:1292–6
Sandford AJ, Shirakawa T, Moffatt MF, et al. Localisation of atopy and β subunit of high-affinity IgE receptor (FcεRI) on chromosome 11q. Lancet 1993;341:332–4
Hill MR, James AL, Faux JA, et al. FcεRIβ polymorphism and risk of atopy in a general population sample. Br Med J 1995;311:776–9
Van Der Pouw Kraan TC, Van Veen A, Boeije LC, et al. An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun 1999;1:61–5
Noguchin E, Nukaga-Nishio Y, Yokoushi Y, et al. Haplotypes of the 5′ region of the IL-4 gene and SNPs in the intergene sequence between the IL-4 and IL-13 genes are associated with atopic asthma. Human Immunol 2001;62:1251–7
Acknowledgments
I thank Ann-Margreth Carlsson for skilful technical assistance. The studies was supported by grants from The Medical Faculty of the University of Lund and the University Hospital of Lund, The Swedish Asthma and Allergy Association and the Vardal Foundation, Sweden.
Author information
Authors and Affiliations
Corresponding author
Additional information
Presented at the First Robert A Good Society Symposium, St. Petersburg, FL 2006.
Rights and permissions
About this article
Cite this article
Oxelius, VA. Immunoglobulin constant heavy G subclass chain genes in asthma and allergy. Immunol Res 40, 179–191 (2008). https://doi.org/10.1007/s12026-007-0007-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-007-0007-1