Abstract
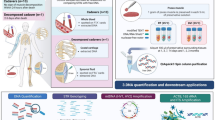
When a firearm projectile hits a biological target a spray of biological material (e.g., blood and tissue fragments) can be propelled from the entrance wound back towards the firearm. This phenomenon has become known as “backspatter” and if caused by contact shots or shots from short distances traces of backspatter may reach, consolidate on, and be recovered from, the inside surfaces of the firearm. Thus, a comprehensive investigation of firearm-related crimes must not only comprise of wound ballistic assessment but also backspatter analysis, and may even take into account potential correlations between these emergences. The aim of the present study was to evaluate and expand the applicability of the “triple contrast” method by probing its compatibility with forensic analysis of nuclear and mitochondrial DNA and the simultaneous investigation of co-extracted mRNA and miRNA from backspatter collected from internal components of different types of firearms after experimental shootings. We demonstrate that “triple contrast” stained biological samples collected from the inside surfaces of firearms are amenable to forensic co-analysis of DNA and RNA and permit sequence analysis of the entire mtDNA displacement-loop, even for “low template” DNA amounts that preclude standard short tandem repeat DNA analysis. Our findings underscore the “triple contrast” method’s usefulness as a research tool in experimental forensic ballistics.

Similar content being viewed by others
References
Gross H. Handbuch für Untersuchungsrichter, Polizeibeamte, Gendarmen u.s.w. 2nd ed. Graz: Leuschner & Lubinsky; 1894.
Weimann W. Über das Verspritzen von Gewebsteilen aus Einschussöffnungen und seine kriminalistische Bedeutung. Dtsch Z Gerichtl Med. 1931;17:92–105.
Brüning A, Wiethold F. Die Untersuchung und Beurteilung von Selbstmörderschusswaffen. Dtsch Z Gerichtl Med. 1934;23:71–82.
Stone IC. Observations and statistics relating to suicide weapons. J Forensic Sci. 1987;32:711–6.
Stone IC. Characteristics of firearms and gunshot wounds as markers of suicide. Am J Forensic Med Pathol. 1992;13:275–80.
Visser JM. Detection and significance of blood in firearms used in contact gunshot wounds. Dissertation: University of Pretoria; 2003.
Schyma C, Madea B, Courts C. Persistence of biological traces in gun barrels after fatal contact shots. Forensic Sci Int Genet. 2013;7:22–7.
Courts C, Gahr B, Madea B, Schyma C. Persistence of biological traces at inside parts of a firearm from a case of multiple familial homicide. J Forensic Sci. 2014;59:1129–32.
Lux C, Schyma C, Madea B, Courts C. Identification of gunshots to the head by detection of RNA in backspatter primarily expressed in brain tissue. Forensic Sci Int. 2014;237:62–9.
Schyma C, Lux C, Madea B, Courts C. The ‘triple contrast’ method in experimental wound ballistics and backspatter analysis. Int J Legal Med. 2015;. doi:10.1007/s00414-015-1151-0.
Eichmann C, Parson W. ‘Mitominis’: multiplex PCR analysis of reduced size amplicons for compound sequence analysis of the entire mtDNA control region in highly degraded samples. Int J Legal Med. 2008;122:385–8.
Alonso A, Albarran C, Martin P, Garcia P, Garcia O, de la Rua C, et al. Multiplex–PCR of short amplicons for mtDNA sequencing from ancient DNA. Prog Forensic Genet. 2003;9(1239):585–8.
Sijen T. Molecular approaches for forensic cell type identification: on mRNA, miRNA, DNA methylation and microbial markers. Forensic Sci Int Genet. 2014;. doi:10.1016/j.fsigen.2014.11.015.
Grabmüller M, Madea B, Courts C. Comparative evaluation of different extraction and quantification methods for forensic RNA analysis. Forensic Sci Int Gen. 2015;16:195–202.
Pang BC, Cheung BK. Double swab technique for collecting touched evidence. Leg Med (Tokyo). 2007;9:181–4.
Haas C, Hanson E, Bar W, Banemann R, Bento AM, Berti A, et al. mRNA profiling for the identification of blood-Results of a collaborative EDNAP exercise. Forensic Sci Int Genet. 2010;5:21–6.
Sauer E, Madea B, Courts C. An evidence based strategy for normalization of quantitative PCR data from miRNA expression analysis in forensically relevant body fluids. Forensic Sci Int Genet. 2014;11:174–81.
Courts C, Madea B. Specific micro-RNA signatures for the detection of saliva and blood in forensic body-fluid identification. J Forensic Sci. 2011;56:1464–70.
Lindenbergh A, de Pagter M, Ramdayal G, Visser M, Zubakov D, Kayser M, et al. A multiplex (m)RNA-profiling system for the forensic identification of body fluids and contact traces. Forensic Sci Int Genet. 2012;6:565–77.
Wong L, Lee K, Russell I, Chen C. Endogenous controls for real-time quantitation of miRNA using TaqMan microRNA assays. Application Note (Applied Biosystems). 2010:1–8.
Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–11.
Vandesompele J, De Preter K, Pattyn F, Poppe B, van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034.
Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50.
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–15.
Zimmermann P, Bleuler S, Laule O, Martin F, Ivanov NV, Campanoni P, et al. Expression data—a public resource of high quality curated datasets representing gene expression across anatomy, development and experimental conditions. BioData Min. 2014;7:18.
Hruz T, Wyss M, Docquier M, Pfaffl MW, Masanetz S, Borghi L, et al. RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genom. 2011;12:156.
Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3.
Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22.
Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147.
Brüning A. Beiträge zur Untersuchung und Beurteilung von Geschossen usw. Arch Kriminol. 1925;77:85–6.
Betz P, Peschel O, Stiefel D, Eisenmenger W. Frequency of blood spatters on the shooting hand and of conjunctival petechiae following suicidal gunshots wounds to the head. Forensic Sci Int. 1995;76:47–53.
Schyma C. Wounding capacity of muzzle-gas pressure. Int J Legal Med. 2012;126:371–6.
Courts C, Madea B, Schyma C. Persistence of biological traces in gun barrels–an approach to an experimental model. Int J Legal Med. 2012;126:391–7.
Karger B, Nusse R, Schroeder G, Wustenbecker S, Brinkmann B. Backspatter from experimental close-range shots to the head I. Macrobackspatter. Int J Legal Med. 1996;109:66–74.
Karger B, Nusse R, Troger HD, Brinkmann B. Backspatter from experimental close-range shots to the head. II. Microbackspatter and the morphology of bloodstains. Int J Legal Med. 1997;110:27–30.
Karger B, Nusse R, Bajanowski T. Backspatter on the firearm and hand in experimental close-range gunshots to the head. Am J Forensic Med Pathol. 2002;23:211–3.
Kunz SN, Brandtner H, Meyer HJ. Characteristics of backspatter on the firearm and shooting hand—an experimental analysis of close-range gunshots. J Forensic Sci. 2015;60:166–70.
Budowle B. Mitochondrial DNA: a possible genetic material suitable for forensic analysis. In: Lee HC, Gaensslen RE, editors. Advances in forensic science. Chicago: Year Book Medical Publishers; 1990. p. 76–97.
Berger C, Parson W. Mini-midi-mito: adapting the amplification and sequencing strategy of mtDNA to the degradation state of crime scene samples. Forensic Sci Int Genet. 2009;3:149–53.
Phang TW, Shi CY, Chia JN, Ong CN. Amplification of cDNA via RT-PCR using RNA extracted from postmortem tissues. J Forensic Sci. 1994;39:1275–9.
Bauer M, Kraus A, Patzelt D. Detection of epithelial cells in dried blood stains by reverse transcriptase–polymerase chain reaction. J Forensic Sci. 1999;44:1232–6.
Juusola J, Ballantyne J. Messenger RNA profiling: a prototype method to supplant conventional methods for body fluid identification. Forensic Sci Int. 2003;135:85–96.
Bauer M, Gramlich I, Polzin S, Patzelt D. Quantification of mRNA degradation as possible indicator of postmortem interval—a pilot study. Leg Med (Tokyo). 2003;5:220–7.
Miller CL, Diglisic S, Leister F, Webster M, Yolken RH. Evaluating RNA status for RT-PCR in extracts of postmortem human brain tissue. Biotechniques. 2004;36:628–33.
van den Berge M, Carracedo A, Gomes I, Graham EA, Haas C, Hjort B, et al. A collaborative European exercise on mRNA-based body fluid/skin typing and interpretation of DNA and RNA results. Forensic Sci Int Genet. 2014;10:40–8.
Lindenbergh A, Maaskant P, Sijen T. Implementation of RNA profiling in forensic casework. Forensic Sci Int Genet. 2013;7:159–66.
Hanson EK, Lubenow H, Ballantyne J. Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal Biochem. 2009;387:303–14.
Zubakov D, Boersma AW, Choi Y, van Kuijk PF, Wiemer EA, Kayser M. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int J Legal Med. 2010;124:217–26.
Courts C, Madea B. Micro-RNA—a potential for forensic science? Forensic Sci Int. 2010;203:106–11.
Wang Z, Zhang J, Luo H, Ye Y, Yan J, Hou Y. Screening and confirmation of microRNA markers for forensic body fluid identification. Forensic Sci Int Genet. 2013;7:116–23.
Acknowledgments
The authors would like to thank the DFG (Deutsche Forschungsgemeinschaft) and the SNF (Swiss National Science Foundation) for funding this project. The expert technical assistance of Julia Brünig and Marion Sauer is also gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
12024_2015_9695_MOESM1_ESM.tif
Selection of a reference gene for mRNA expression data normalization using the “RefGenes” online resource (TIFF 1469 kb)
12024_2015_9695_MOESM2_ESM.tif
Imaging of PCR products resulting from two multiplex PCRs using “Mito-Mini” primer sets in “low template” samples. PCR products were separated via microfluid-gelelectrophoresis on an Agilent 2100 Bioanalyzer; Primer: primer name; bp: base pairs (TIFF 1320 kb)
Rights and permissions
About this article
Cite this article
Grabmüller, M., Schyma, C., Euteneuer, J. et al. Simultaneous analysis of nuclear and mitochondrial DNA, mRNA and miRNA from backspatter from inside parts of firearms generated by shots at “triple contrast” doped ballistic models. Forensic Sci Med Pathol 11, 365–375 (2015). https://doi.org/10.1007/s12024-015-9695-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12024-015-9695-3