Abstract
Grading of gastrointestinal neuroendocrine neoplasms (GI-NENs) relies mainly on mitotic activity and Ki-67 proliferation index. It is often difficult to predict metastatic potential of these neoplasms. Recent studies have shown that GI-NENs express a wide spectrum of microRNAs. We examined two microRNAs (miR-96 and miR-133a) that were recently identified in GI-NENs to determine if they could assist in evaluating the biological behavior of these neoplasms. A tissue microarray (TMA) was constructed with 51 primary GI-NENs, mainly from the small intestine and metastatic tumors from the same cases, including liver metastases (N = 20) and lymph node metastases (N = 33). The cases were immunohistochemically stained for chromogranin A, synaptophysin, and Ki-67. In situ hybridization (ISH) was done with probes from Exiqon (Woburn, MA). Quantitative RT-PCR (qRT-PCR) was also performed on all the cases (N = 105). ISH analysis showed that miR-96 expression was significantly higher in the liver metastatic neoplasms compared to the primary NENs (p < 0.05); however, it was not significant for miR-133a expression levels. qRT-PCR showed that miR-96 levels were increased during progression from the primary tumors to metastases in the liver. qRT-PCR showed a decrease in miR-133a in the liver metastases compared to the primary tumors (p < 0.05). Appendiceal carcinoids without metastases (n = 3) had low levels of miR-96 and high levels of miR-133a by qPCR. The study suggests that analysis of these two microRNAs by qRT-PCR may be useful in detecting more aggressive GI-NENs and that ISH analysis may also assist in the evaluation of patients with GI-NENs.
Similar content being viewed by others
Introduction
The incidence of gastrointestinal neuroendocrine neoplasms (GI-NENs) has been increasing [1]; however, the pathogenesis of these neoplasms remains unknown. Patients may present with advanced disease with metastases to liver, lymph nodes, and other sites, and these metastases are sometimes unresectable. There is no definite curative therapy for metastatic GI-NENs [2]. Grading of GI-NENs relies mainly on mitotic activity and Ki-67 proliferation index. It is often difficult to predict metastatic potential in these neoplasms. There is increasing understanding that they have a different pathogenesis from pancreatic neuroendocrine tumors.
MicroRNAs (miRNAs), a family of 21 to 25 nucleotide sequence, are noncoding small ribonucleic acids that are implicated in the regulation of various hosts of cellular processes [3]. The first identified miRNA, lin-4 in Caenorhabditis elegans by Ambros et al., was shown to have important roles in controlling timing of larval development through regulation of messenger RNA (mRNA) translation [4]. Since then, more than 1700 miRNAs have been discovered, and it is estimated to be more than 2500 miRNA in the human genome. miRNAs function at post-transcriptional level in embryonic development or in stem cell self-renewal, and they may act as oncogenes and tumor suppressors. miRNAs are more specific compared to mRNA in classification of cancers [5]. They are more stable in a cell-free environment and do not easily degrade in unfavorable physiological conditions such as extreme pH or temperature variation [6]. Their differential expression has been studied for diagnostic purposes as well as for therapeutic targets [7, 8].
Since each miRNA is potentially able to target a large number of genes involved in cell processes through imprecise binding to mRNA, the challenging task is to elucidate their value in the initiation and development of tumorigenesis.
Many studies have now revealed that different cancer types have distinctive miRNA expression signatures [9]. Several studies have already reported specific miRNA biomarkers in different types of cancer such as hematological cancer [10, 11], lung cancer [12], prostate cancer [13], pancreatic cancer [14], liver cancer [15], colorectal cancer [16], gastric cancer [17], ovarian cancer [18], esophageal cancer [19], cervical cancer [20], thyroid cancer [21], astrocytomas [22], head and neck cancer [23], and bladder cancer [23]. There are emerging studies on the use of specific miRNA signatures to predict clinical outcomes of lung neuroendocrine tumors and GI-NENs; the latter, however, was an animal model [24]. There are limited studies on miRNA expression patterns and clinical outcome studies on human GI-NENs. A study by Ruebel et al. showed differential expression of miRNAs in GI-NENs which had downregulation of miRNA-133a, miRNA-145, miRNA-146, miRNA-222, and miRNA-10b in metastatic compared to primary tumors and upregulation of miRNA-183, miRNA-488, and miRNA-19a+b in metastatic tumors [25]. A follow-up study by Li et al. showed downregulation of miR-133a-31, miR-129-5p, and miR-215 and upregulation of miR-96, miR-182, miR-183, miR-196a, and miR-200a [26]. Recent studies suggest that miRNA detection in liquid biopsies may have good specificity and sensitivity, is cost-effective, and offers superior noninvasive markers [27, 28], so this may be a practical approach to evaluate tumor progression. We examined two microRNAs (miR-96 and miR-133a) that were recently identified in GI-NENs to determine if they could assist in evaluating the biological behavior of GI-NENs.
Materials and Methods
Tissue Samples
Three groups of 51 primary GI-NENs were designated as shown in Table 1. The first group included 19 cases with primary tumors from small intestine with lymph node metastasis (N = 19). The second group included 21 cases with primary tumors from small intestine with liver metastasis (N = 21) and lymph node metastasis (N = 14). The third group included 11 cases with only primary tumor (8 cases from small intestine and 3 cases from appendix) and no known distant metastasis. None of the group 3 cases revealed lymphovascular invasion.
A tissue microarray (TMA) was constructed with 51 primary GI-NENs mainly from the small intestine and metastatic tumors from the same cases including liver metastases (LVM, N = 21) and lymph node metastases (LNM, N = 33). Immunohistochemistry was performed on TMA section for chromogranin A, synaptophysin, and Ki-67. In situ hybridization (ISH) was performed on the TMA with probes from Exiqon (Woburn, MA) as previously reported [29]. Staining on the TMA was evaluated using a scale of 0 to 3+ for the intensity of staining and the percentage of tumor cells showing positive staining. Permission for the study was approved by the institutional review board committee of University of Wisconsin.
In Situ Hybridization
Formalin-fixed paraffin-embedded (FFPE) TMA sections with probes labeled at both 3′ and 5′ ends with a double digoxigenin (DIG)-labeled locked nucleic acid (LNA) were used for ISH. LNA ISH on paraffin tissue sections with probe specific for human miR-133a (614419-360; Exiqon, Woburn, MA) and miR-96 (614085-360; Exiqon, Woburn, MA) was performed according to the manufacturer’s instructions (Exiqon, Woburn, MA) as previously reported [29]. In brief, 4-μm sections of archived paraffin-embedded specimens were deparaffinized in xylenes and then rehydrated through an ethanol dilution series (from 100 to 70%). Sections were treated with proteinase K at 37 °C for 10 min and then dehydrated through an ethanol dilution series (from 70 to 100%). DIG-labeled probes were denatured at 94 °C for 5 min. Slides were incubated with the probes diluted to 250 nM in hybridization buffer at 55 °C for 2 h. Stringent washes were performed with 5× saline sodium citrate (SSC), 1× SSC, and 0.2× SSC buffers at 50 °C 33 min; DIG blocking reagent (Roche, Indianapolis, IN) in maleic acid buffer containing 2% sheep serum at 30 °C for 15 min; alkaline phosphatase-conjugated anti-digoxigenin (diluted 1:800 in blocking reagent, Roche, Indianapolis, IN) at room temperature for 60 min; enzymatic development using 4-nitro-blue tetrazolium (NBT) and 5-brom-4-chloro-30-indolyl-phosphate (BCIP) substrate (Vector Laboratories, Burlingame, CA) developing at 30 °C for 120 min, followed by nuclear fast counterstain for 1 min. The slides were then placed in water, dehydrated in alcohol and xylene, and mounted with mounting medium. Scrambled probe (699004-360; Exiqon, Woburn, MA) was used as a negative control, while U6 (699002-360; Exiqon, Woburn, MA) served as positive control. ISH scoring was performed by two independent observers using conventional bright field microscopy, and differences in interpretation were reviewed for consensus. Cytoplasmic staining was interpreted based on the intensity as negative (0), weak (1+), moderate (2+), and strong (3+). Cases showing 1+, 2+, and 3+ intensity of staining were considered as positive. The expression was considered focal when 1–25% of cells were positive and diffuse when > 25% of cells were positive. Staining of < 1% of all tumor cells was considered to be negative.
RNA Extraction and qRT-PCR
Total RNA was extracted from FFPE samples using the Maxwell® 16 LEV RNA FFPE Purification Kit (AS1260, Promega, Madison, WI) according to the manufacturer’s instructions. cDNA was constructed and qPCR performed using the GeneCopoeia All-in-One™ miRNA Quantitative RT-PCR (qRT-PCR) Detection Kit (SYBR Green; QP016; GeneCopoeia, Rockville, MD) according to the manufacturer’s instructions on the Roche LightCycler 2.0 (Roche, Indianapolis, IN). Primers used were miR-96 F 5′-CGGCGGTTTGGCACTAGCACATT-3′ and miR-133a F-5′-TTTGGTCCCCTTCAACCAGCTG-3′. The miRNA kit provides a universal reverse primer. Results were normalized to RN5-8S1 (HmiRQP9031, GeneCopoeia, Rockville, MD).
qRT-PCR was also performed on 105 tissue blocks.
Statistical Analysis
All qRT-PCR analyses were performed in duplicates. The statistical significance of the difference between three groups was evaluated by one-way ANOVA followed by Bonferroni test. p value < 0.05 is considered significant.
Results
There were more female patients than male (63 versus 37%) with GI-NENs. The age of patients ranged from 17 to 80 years old. Among group 1 (primary tumor with lymph node metastasis), 17 of 19 (89.5%) patients were alive at the time of data collection, 18 of 21 (85.7%) among group 2 and 10 of 11 (90.9%) among group 3 patients (Table 1). The cause of death in one of the patients among group 3 was renal transplant-associated complications. Tumor size varied from 0.45 to 10.0 cm among groups 1 and 2 and 0.2 to 4.0 cm among group 3 patients. The majority of cases were grade 1–2 neoplasms with Ki-67 proliferation index ranging from less than 1 to 20% with the exception of one case where the Ki-67 index was 90%.
Immunohistochemical (IHC) staining of the TMA for Ki-67 showed that the majority of the NENs had a low Ki-67 proliferation index except for one case in group 2 where the Ki-67 index was 90%. Based on Ki-67 per diagnosis, 98% of cases (50 out of 51) were either grade I or II tumors. Chromogranin and synaptophysin IHC showed positive staining in all of the primary and metastatic neoplasms.
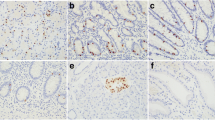
ISH analysis showed predominantly cytoplasmic expression of miR-133a and miR-96. Positive controls with U6 showed cytoplasmic and nuclear staining, while the negative controls resulted in no staining on the slides. miR-96 expression was significantly higher in the liver metastatic neoplasms compared to the primary NENs (p < 0.05) (Fig. 1a–c). ISH analysis of miR-133a showed a decreasing trend of expression of this microRNA in the liver and lymph node metastases compared to the primary tumors, but the differences did not reach significance. Expression of miR-96 and miR-133a was not associated with tumor size.
In situ hybridization analysis of miR-96 and miR-133 expression in gastrointestinal neuroendocrine neoplasms (GI-NENs). a In situ hybridization of primary tumor with mild 1+ expression of miR-96 and the enterochromaffin cells in adjacent normal gastrointestinal mucosa (upper half) with 1+ expression. b Liver metastasis with strong 3+ expression of miR-96. c Lymph node metastasis with strong 3+ expression of miR-96. d In situ hybridization of primary tumor with strong 3+ expression of miR-133. e Liver metastasis with mild 1+ expression of miR-133 and adjacent normal liver (right half). f Lymph node (LN) metastasis with mild 1+ expression of miR-133
qRT-PCR showed that miR-96 levels were increased during progression from the primary tumors to metastases in the liver (Figs. 2 and 3a). qRT-PCR showed a significant decrease in miR-133a in the liver metastases compared to the primary tumors (p < 0.05) (Fig. 3b).
In situ hybridization scoring of miR-96 was performed on a scale of 0 to 3+ as described in “Materials and Methods” section. The chart shows significant increase at metastatic liver (p < 0.05). LN lymph node metastasis
Appendiceal carcinoids (n = 3) without metastases had low levels of miR-96 and high levels of miR-133a by qRT-PCR.
Discussion
Analysis of miRNA expression by in situ hybridization and qRT-PCR showed that GI-NENs express miR-96 and miR-133a in both the primary and metastatic tumor sites in the lymph nodes and the liver. There was increased expression of miR-96 and decreased expression of miR-133a during progression from primary to metastatic GI-NENs. This is a larger study where we focused on two miRNAs which were previously shown to be expressed in GI-NENs. These findings are consistent with reports from our earlier studies [25, 26]. The results suggest that these miRNA expression levels have an important role in tumor progression of GI-NENs. miR-133a is located on chromosome 18 and has also been studied in skeletal and cardiac muscle, whereas miR-96 is located on the long arm of chromosome 7 at position 32.2.
The majority of GI-NENs arise in the distal jejunum and ileum and commonly metastasize to the liver and lymph nodes. Tumor size does not reliably predict metastatic potential. Metastases were also found in patients with primary tumors smaller than 1 cm in diameter. This led to some difficulty in gathering control cases for group 3 cases. Markers commonly used to follow patients after surgery include chromogranin A and chromogranin B which are relatively specific compared to other markers which may be elevated in GI-NENs such as pancreatic polypeptide, neuron-specific enolase, chorionic gonadotropin α chain, and chorionic gonadotrophin β chain. Although chromogranin A and chromogranin B are more specific markers, these can be elevated in other conditions such as patients with renal failure and enterochromaffin cell hyperplasia secondary to the use of proton pump inhibitors. Although a few NET markers exist, there is a paucity of sensitive and specific marker to predict tumor growth and behavior. Because miRNAs are relatively stable molecules in a cell-free environment as well as in paraffin-embedded tissue, these may serve as new sensitive tissue markers. The level of increase of miR-96 and decrease of miR-133a by qRT-PCR in metastatic disease is quite variable, but a combination of a biomarker that increases with tumor progression with another biomarker that decreases during tumor progression is a reasonable strategy to detect changes in patients with residual or recurrent NETs. These results suggest that analysis of these two microRNAs by qRT-PCR may be useful in detecting metastatic disease in liquid biopsy specimens and that ISH analysis may also assist in the evaluation of patients with GI-NENs if biopsy tissues are available. Furthermore, since there are no reliable methods to predict which NETs will respond to aggressive chemotherapy, the result of differential miRNA expression may serve as molecular therapeutic targets.
Currently, most of the biomarkers are based on proteins that exist in serum/plasma. There are major limitations of protein-based markers such as low sensitivity and specificity with false positive or false negative results. miRNA expression is tissue or tumor specific, and the shorter length of miRNAs makes them less susceptible to degradation by chemicals and physical environment, and they are a stable cell-free biomarker. miRNAs are ubiquitous in many body fluids such as saliva, serum, blood, urine, breast milk, and even cerebrospinal fluid [30]. The mechanism by which miRNA escapes degradation by RNAses in body fluids is not known. The remarkable stability of miRNA provides great advantage for their use as diagnostic tools in liquid biopsy specimens in oncology [27].
The use of miRNA as a diagnostic tool is encouraging; however, there are still technical and theoretical challenges that need to be addressed. For example, there is a need for learning more about the oncogenic mechanisms of miRNA expression. There is a need for refining the data of tissue-specific miRNA expression using larger samples and excluding potential cross-reactivity between different microRNAs. miR-133a expression is not specific for GI-NENs. This microRNA has also been reported in astrocytomas and hepatocellular carcinomas, and in patients with acute myocardial infarction [31]. miR-96 has also been associated with other malignancies such as nonsmall cell lung cancer and breast cancer [32, 33]. Thus, a combination of various microRNAs would increase the specificity of diagnostic tests for GI-NETs.
In summary, we have used a combination of in situ hybridization on formalin-fixed paraffin-embedded tissues and qRT-PCR to analyze expression of miR-96 and miR-133a in primary and metastatic GI-NETs. This suggests that a combination of miR-96 and miR-133a may serve as useful diagnostic and prognostic markers for GI-NENs.
References
Modlin IM, Oberg K, Chung DC, Jensen RT, de Herder WW, Thakker RV, et al. Gastroenteropancreatic neuroendocrine tumours. Lancet Oncol 9:61–72, 2008.
Cunningham JL, Diaz de Stahl T, Sjoblom T, Westin G, Dumanski JP, Janson ET. Common pathogenetic mechanism involving human chromosome 18 in familial and sporadic ileal carcinoid tumors. Genes Chromosomes Cancer 50:82–94, 2011.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297, 2004.
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854, 1993.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature 435:834–838, 2005.
Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS One 3, 2008.
Debernardi S, Massat NJ, Radon TP, Sangaralingam A, Banissi A, Ennis DP, et al. Noninvasive urinary miRNA biomarkers for early detection of pancreatic adenocarcinoma. Am J Cancer Res 5:3455–3466, 2015.
Li Y, Sarkar FH. MicroRNA Targeted Therapeutic Approach for Pancreatic Cancer. Int J Biol Sci 12:326–337, 2016.
Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet 10:578–585, 2009.
Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol, 141:672–675, 2008.
Fayyad-Kazan H, Bitar N, Najar M, Lewalle P, Fayyad-Kazan M, Badran R, et al. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J Transl Med, 11:31, 2013.
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res, 18:997–1006, 2008.
Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer, 128:608–616, 2011.
Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA, 311:392–404, 2014.
Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol, 45: 355–360, 2011.
Ayaz L, Görür A, Yaroglu HY, Ozcan C, Tamer L. Differential expression of microRNAs in plasma of patients with laryngeal squamous cell carcinoma: potential early-detection markers for laryngeal squamous cell carcinoma.J Cancer Res Clin Oncol 139:1499–1506, 2013.
Tsujiura M., Komatsu S, Ichikawa D, Shiozaki A, Konishi H, Takeshita H, et al. Circulating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancer. Gastric Cancer, 18:271–279, 2015.
Zheng H, Zhang L, Zhao Y, Yang D, Song F, Wen Y, et al. Plasma miRNAs as diagnostic and prognostic biomarkers for ovarian cancer. PLoS One, 8, 2013.
Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, et al Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer, 105:104–111, 2011.
Yu J, Wang Y, Dong R, Huang X, Ding S, Qiu H. Circulating microRNA-218 was reduced in cervical cancer and correlated with tumor invasion. J Cancer Res Clin Oncol, 138:671–674, 2012.
Yu S, Liu Y, Wang J, Guo Z, Zhang Q, Yu F, et al. Circulating microRNA profiles as potential biomarkers for diagnosis of papillary thyroid carcinoma. J Clin Endocrinol Metab, 97:2084–2092, 2012.
Yang C, Wang C, Chen X, Chen S, Zhang Y, Zhi F, et al. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer, 132:116–127, 2013.
Snowdon J, Boag S, Feilotter H, Izard J, Siemens DR. A pilot study of urinary microRNA as a biomarker for urothelial cancer. Can Urol Assoc J, 7:28–32, 2013.
Gao Y, Schug J, McKenna LB, Le Lay J, Kaestner KH, Greenbaum LE. Tissue-specific regulation of mouse microRNA genes in endoderm-derived tissues. Nucleic Acids Res 39:454–463, 2011.
Ruebel K, Leontovich AA, Stilling GA, Zhang S, Righi A, Jin L, et al. MicroRNA expression in ileal carcinoid tumors: downregulation of microRNA-133a with tumor progression. Mod Pathol 23:367–375, 2010.
Li SC, Essaghir A, Martijn C, Lloyd RV, Demoulin JB, Oberg K, Giandomenico V. Global microRNA profiling of well-differentiated small intestinal neuroendocrine tumors. Mod Pathol 26:685–696, 2013.
Izzotti A, Carozzo S, Pulliero A, Zhabayeva D, Ravetti JL, Bersimbaev R. Extracellular MicroRNA in liquid biopsy: applicability in cancer diagnosis and prevention. Am J Cancer Res 6:1461–1493, 2016.
Shigeyasu K, Toden S, Zumwalt TJ, Okugawa Y, Goel A. Emerging Role of MicroRNAs as Liquid Biopsy Biomarkers in Gastrointestinal Cancers. Clin Cancer Res 23:2391–2399, 2017.
Guo Z, Hardin H, Montemayor-Garcia C, Asioli S, Righi A, Maletta F, et al. In Situ Hybridization Analysis of miR-146b-5p and miR-21 in Thyroid Nodules: Diagnostic Implications. Endocr Pathol 26:157–163, 2015.
Akers JC, Ramakrishnan V, Kim R, Skog J, Nakano I, Pingle S, et al. miR-21 in the Extracellular Vesicles (EVs) of Cerebrospinal Fluid (CSF): A Platform for Glioblastoma Biomarker Development. PLoS One 8(10): e78115, 2013.
He B, Xiao J, Ren A, Zhang Y, Zhang H, Chen M, et al. Role of miR-1 and miR-133a in myocardial ischemic postconditioning. Journal of Biomedical Science 18:22, 2011.
Cai T, Long J, Wang H, Liu W, Zhang Y. Identification and characterization of miR-96, a potential biomarker of NSCLC, through bioinformatic analysis. Oncol Rep 38:1213–1223, 2017.
Moazzeni H, Najafi A, Khani M. Identification of direct target genes of miR-7, miR-9, miR-96, and miR-182 in the human breast cancer cell lines MCF-7 and MDA-MB-231. Mol Cell Probes 34:45–52, 2017.
Funding
This research project was supported by the Department of Pathology Research Fund from the University of Wisconsin School of Medicine and Public Health (to RM).
The authors thank the University of Wisconsin Translational Research Initiatives in Pathology laboratory, in part supported by the UW Department of Pathology and Laboratory Medicine and UWCCC grant P30 CA014520-39, for use of its facilities and services.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Mandal, R., Hardin, H., Baus, R. et al. Analysis of miR-96 and miR-133a Expression in Gastrointestinal Neuroendocrine Neoplasms. Endocr Pathol 28, 345–350 (2017). https://doi.org/10.1007/s12022-017-9504-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12022-017-9504-5