Abstract
Textpresso is a text-mining system for scientific literature. Its two major features are access to the full text of research papers and the development and use of categories of biological concepts as well as categories that describe or relate objects. A search engine enables the user to search for one or a combination of these categories and/or keywords within an entire literature. Here we describe Textpresso for Neuroscience, part of the core Neuroscience Information Framework (NIF). The Textpresso site currently consists of 67,500 full text papers and 131,300 abstracts. We show that using categories in literature can make a pure keyword query more refined and meaningful. We also show how semantic queries can be formulated with categories only. We explain the build and content of the database and describe the main features of the web pages and the advanced search options. We also give detailed illustrations of the web service developed to provide programmatic access to Textpresso. This web service is used by the NIF interface to access Textpresso. The standalone website of Textpresso for Neuroscience can be accessed at http://www.textpresso.org/neuroscience/.
Similar content being viewed by others
Introduction
Literature is an important and fundamental element of the scientific realm and plays a central role of communication among researchers, from the exchange of latest findings and dispersion of thoughts and discussions, to the detailed description of experiments. At the same time, the size and growth rate of scientific literature have made it nearly impossible for the researcher to keep up with articles relevant to his or her area of interest; for example, PubMed now comprises 17 million citations and adds 700,000 entries every year. There is therefore a need for computational approaches to filter through the literature and provide the researcher with information specifically relevant to him or her.
Computationally retrieving information from literature is called natural language processing and can be divided into four main areas: information retrieval, information (fact) extraction, document classification and literature-based discovery. Many new and exciting tools and methods in natural language processing have been developed in the past years and are described in Hunter and Cohen (2006) and Zweigenbaum et al. (2007). Information retrieval recovers a pertinent subset of documents. Most such retrieval systems use keywords for searches. Many internet search engines are of this type, e.g., PubMed. Information extraction is the process of obtaining pertinent information (facts) from documents, and this extraction is usually done on a large number of documents. In the context of biological literature, name entity recognition (NER) and the detection or extraction of relationships between entities are major elements of information extraction. Textpresso for Neuroscience has been developed to focus on information retrieval and information extraction.
Common search engines such as Google and Yahoo do not handle scientific literature as well as one might like even with specialization such as Google Scholar. Keywords—defined as words used in some manner by a search engine to identify relevant documents—are usually found within a document, but when typing in a set of words, one often wants the search scope to be within a sentence, as interesting facts or biological data are often (but not always) expressed in one or a few sentences. Thus, Textpresso defines keywords as tokens found in documents and indexed for lookup on a sentence level. In addition, most search engines allow only the abstract of a paper to be searched, and much information important to the scientist is therefore lost because it is buried in the full text. Full text contains redundancies, which increase the chances of obtaining a hit with a query using a pure keyword search engine. However, if full text is used, but a search engine is not restricted to a particular scientific literature, search returns are heavily diluted with false positives (hits that are irrelevant or incorrect).
Pure keyword search engines have another drawback: let us assume a researcher is eager to resurrect a certain fact he vaguely knows about. He tries to find it by typing a carefully crafted set of keywords but fails to succeed. He then tries to refine his query by adding more keywords, resulting in fewer and fewer returns, until he ends up with none. Furthermore, neither general nor more semantic questions can be answered with pure keyword engines. Consider the query: “Which gene interacts with my favorite gene X?” With a keyword engine, gene name X and names of other genes suspected of being an interaction candidate would have to be typed in; moreover, some words that mean ‘interaction’ would be necessary to filter the results, since two genes could be mentioned in other context, such as in a list of genes in microarray results.
The latter situation can be significantly alleviated through the introduction of semantic categories or concepts (we will use the terms interchangeably throughout this paper). A category is a bag of words and phrases that have a common meaning, and usually the category is named after the meaning that groups them together. If we fill this bag—which we call a lexicon in this article—with all terms in the domain known to be relevant and true, mark up and index the whole corpus with all occurrences of terms of the category, then a query that includes searching for these instances in the text is bound to be much more efficient. In the example above, the query would consist of the keyword X together with the categories gene and interaction. The interaction category contains words such as ‘bind’, ‘interact’, ‘attach’ and ‘suppress’ (and their lexical variations) as well as the corresponding nouns. The gene category holds terms such as ‘locus’, the word ‘gene’ itself as well as specific gene names such as ‘wingless’, ‘let-60’, ‘TP53’, which usually comprise the majority of entries in the gene lexicon. Thus, lexica are the vocabularies of their corresponding categories. Synonyms are included in categories, and a set of synonyms can constitute a category, but a category usually contains more than synonyms. The verb ‘heighten’ might be considered a synonym for ‘enhance’, but is certainly not a synonym of ‘silence’. However, all three verbs can be considered members of a regulation category.
When designing Textpresso, we took all these considerations into account: we wanted to build a search engine that is focused on particular biological literatures, searches the full text of research articles, and, besides keyword searches, allows for searching instances of semantic categories, as we believe that it adds meaning to a query. Textpresso is meant to be a practical tool for researchers and biological database curators, and at the same time a platform for natural language processing to help extract information on a massive scale.
Textpresso for Neuroscience is part of the Neuroscience Information Framework (Gardner et al. 2008), which can be accessed through http://nif.nih.gov. It is a platform that enables the Neuroscience community to locate and query online resources relevant to Neuroscience; key features of NIF are the capability to register these resources at various levels of depth, from simply providing a URL with a sparse description of a resource, to sophisticated annotations of single data items across databases, that can be queried based on concepts. These concepts are organized through a structured, controlled vocabulary.
General Features of Textpresso
Textpresso has two central features: first it offers search access to individual sentences of full text papers, and second, it introduces categories, marks up the instances of the categories in the corpus of literature and allows the user to search for instances of these categories in the full text (Müller et al., 2004). Categories can be: (1) biological concepts (e.g., receptor, brain area, cellular component), (2) relationships between two or more objects such as association and regulation, and (3) descriptions (e.g., method, characterization). Accordingly a search query can be formulated that may consist of keywords, phrases and the selection of one or more categories (at least one item must be present). A category hit occurs when a particular word or phrase in the sentence is defined as a member of a lexicon of the particular category; a keyword or phrase hit occurs when the particular keyword or phrase itself is present as a token or a sequence of tokens in the sentence. Textpresso searches for these query items in sentences, and returns sentences that contain all of them in one sentence. Assuming that many facts are expressed in one sentence, such a search can be very powerful and retrieve facts of interest: for example, if one wants to find out with which neural cell types in specific brain areas the TRP channel TRPC1 is associated, one enters the keyword TRPC1 and selects the two categories NIF (neural) cell types and brain area. Even though a semantically complete question was the starting point of the query, the query itself only contains one keyword and two categories chosen from a menu. Sentences containing all three query items are displayed in the return, and the likelihood of a significant search return is increased as the specification of categories as semantic concepts adds meaning to a query. In particular, a search of 67,500 papers for TRPC1 returns 716 sentences in 96 papers; a search for TRPC1 and neural cell type returns 18 sentences in ten papers, while demanding TRPC1 and both categories returns eight sentences in four papers. By contrast, neural cell types are mentioned in 305,409 sentences in 37,959 papers. If we replace the keyword TRPC1 with the category TRP channels, 18 sentences in eight papers are returned. In this query we asked the search engine to return all TRP channels that are mentioned in connection with neural cell types and brain area. This query and its result are displayed in Fig. 1.
Example of complex query without any keywords: What TRP channels are associated with particular neural cell types in specific brain areas? No keywords are used but three categories. 18 sentences are identified in 8 papers from 67,500 papers. Note that this query returns more hits than when replacing the category ‘TRP channel’ with the keyword ‘TRPC1’ as the category comprises more terms than just one keyword
Another, more complex example is illustrated in Fig. 2. In this case the researcher is interested in whether any prescription drugs of abuse other than nicotine are associated with nicotinic receptors. We include the keywords nicotinic receptor but exclude nicotine, by preceding it with a minus (−) sign, and choose the category Prescription Drugs of Abuse. 150 matches in 79 documents from 67,500 papers are returned.
Textpresso was originally developed for C. elegans literature, but search engines for many other literatures have now been deployed. All literatures share a core set of categories, and in addition to them, categories specific to the particular literature are implemented. Each category comes with a corresponding lexicon which is filled with thousands of words and phrases. We obtain these words and phrases from ontologies such as the Gene Ontology (The Gene Ontology Consortium, 2000, 2008). All three major GO categories—molecular function, biological process and cellular component—and their first children are part of the core categories. Further sources for the lexica are model organism databases, from which we mostly obtain lists of biological entities such as gene names, anatomies and phenotypes. We have done this in the past for our Drosophila, Arabidopsis and C. elegans sites.
As of April 2008, nineteen Textpresso systems have been deployed worldwide, comprising approximately 65 million sentences in 190,000 full text papers. We maintain four sites, the C. elegans site with 11,500 full text papers (in collaboration with WormBase), the Drosophila site with 20,100 papers (in collaboration with FlyBase), the Arabidopsis site with 15,100 papers (in collaboration with The Arabidopsis Information Resource) and the Neuroscience system with 67,500 full text papers (as part of the Neuroscience Information Framework).
How does Textpresso compare to some other familiar search engines? PubMed and Google Scholar index more material than does Textpresso for Neuroscience. Google Scholar includes full text but does not use an ontology. PubMed has only abstracts, but does provide some access to information present in full text via manual curation of MeSH terms; keywords entered into PubMed are matched against and mapped onto MeSH terms via an automatic term mapping procedure, and records previously annotated manually with these terms are then retrieved. PubMed organizes MeSH terms and Taxonomy Ids in form of ontologies. The approach of PubMed differs strongly from the strategy Textpresso is pursuing. In the case of Textpresso, all categories and their terms are searched for and mapped onto all full text articles, and subsequently the user can search for occurrences of these categories anywhere in the text, representing a true category search. PubMed queries MeSH terms with keywords, and articles annotated with mapped MeSH terms are retrieved; however, this annotation is much sparser and only applied to the whole document. Neither PubMed nor Google Scholar uses a sentence level scope of query, be it a keyword or category search.
GoPubMed (Doms and Schroeder 2005; http://www.gopubmed.org) analyzes keyword searches submitted to PubMed by matching search results against Gene Ontology and MeSH concepts and terms. The matching is accomplished by using a sophisticated term extraction algorithm based on local sequence alignment of words. GoPubMed then allows browsing and filtering out articles of the original PubMed return that mention matched concepts. Thus, while GoPubMed does not allow category or full text searches, it structures search results in a semantic manner.
Textpresso for Neuroscience
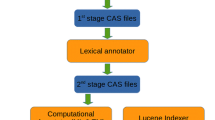
The corpus of the Neuroscience site at http://www.textpresso.org/neuroscience/ is journal-based. We have currently included 18 journals in our corpus which have been selected by researchers and developers of the NIF project based on their perceived importance in the field. We downloaded the bibliographies for all articles in these journals by posting queries to PubMed, using the E-utilities provided by PubMed, by first downloading a list of PMIDs (the unique identifier assigned to a PubMed record), and subsequently retrieving and retaining title, author, year of publication, journal and citation information, and abstract for each PMID. We then obtained the full texts in form of PDFs from the journals. For some journals we only offer searches in abstracts and bibliographies as we did not have a subscription for them. As we needed to be able to convert PDF to plain ASCII text for processing, most articles, for which we obtained a PDF, are from recent years, while older articles, scanned in by publishers as images and transformed into PDFs, could not be included. As these older articles are scanned in as images, they are not text-convertible without further processing, which involves using open character recognition (OCR) software. In some other cases, we could not obtain PDFs for other technical reasons, but we will continue to work on these issues in upcoming database releases. Via a PDF-to-HTML conversion package based on XPDF (an open source software that allows viewing and converting of PDFs), we converted all applicable PDFs first into HTML and then plain ASCII text. The conversion is done through HTML in order to retain formatting information such as italicization which can be used for such tasks as gene identification. The current Neuroscience corpus (as of April 2008) contains 67,500 full text articles, 131,300 abstracts and 148,000 titles available for searching. Some abstracts are missing because they were not provided by PubMed. After populating the Textpresso database with all data, full texts, abstracts and titles were marked up with the Textpresso categories. These markups along with all words in the text were indexed for fast database searches and retrieval.
We have added nine Neuroscience-specific categories to the site to enable the researcher to reduce ambiguities in searches and decrease the rate of false positives for searches. These categories are of high interest to the neuroscientist; they include brain area, drugs of abuse, NICSNP candidate gene, NIF cell type, neuropsychology & behavior, prescription drug of abuse, receptor, substance abuse and TRP channel. Table 1 shows the approximate size of their corresponding lexica together with some example terms. We initiated their implementation after discussion with NIF collaborators. For some categories we downloaded lists of terms from the National Institute on Drug Abuse website (NIDA). Other resources include web pages at the National Center for Biotechnology Information (NCBI) and other publicly available Neuroscience resources on the web. We made lexical variations of all imported terms, which included the capitalized and pluralized versions of them and added them to our lexica. These categories are considered to be a first version and will be refined and expanded further upon consultation with NIF developers.
Textpresso for Neuroscience can be accessed in two ways. A web interface enables the user to interactively search the literature, while web services allow access in an automated fashion making it possible to mine the literature via scripts and programs. Both access modes are utilized by NIF.
The homepage consists of the search interface, a description of the current database as well as a News and Messages section. The text field of the search interfaces allows for entering keywords and phrases. Phrases have to be put in double quotes. White spaces between keywords or phrases act as the Boolean operation AND. Other Boolean operators available are OR and NOT. A comma indicates that two words or phrases are to be concatenated by an OR. A minus sign (−) indicates that the following keyword or phrase should not appear in the sentence. The checkboxes underneath the text field modify the keyword search. When ‘Exact match’ is clicked, all words have to be matched exactly, while, if it is not clicked, a wild card sign, which represents one or more arbitrary characters, is appended to each word. The checkbox ‘Case sensitive’ controls whether upper and lower case of each word should be considered for the query. The user can furthermore require categories to be added to the queries. Up to four categories can be specified from the cascading menus. They are always concatenated with a Boolean AND. Finally, advanced search options described below can be activated by clicking on the corresponding link.
Pressing the ‘Search!’ button submits the query to the search engine. The user is led to the search interface, and score-sorted list of search returns is returned (Fig. 3). Points are given when a search term is found in a sentence, so that sentences with more matches are given a higher score. On top of the table the user can adjust options to control the global display such as number of entries per page, number of sentences surrounding the matching sentence and search term highlighting. The head of the table displays the number of matches and corresponding documents. Additionally some global links are offered, such as exporting the results in EndNote or XML.
Every document entry contains bibliographical information, abstract, as well as the matching sentences. The matching words and categories are highlighted in the text by default, but this feature can be switched off. Some returned sentences appear to be scrambled due to incorrect conversion from PDF or HTML to text. These are mostly tables and captions. As they are less useful to the user, they are suppressed in the result display, but can be accessed via a special link that opens a new window displaying the scrambled sentence. Particular items such as bibliography or matching sentences in each entry can be collapsed and expanded for clarity of display. Each entry also provides supplemental links, such as a link to the online text, to a list of related articles, to the corresponding PubMed citation, as well as to an export function of the document in EndNote or XML.
Textpresso allows for more advanced search options. This feature is accessed by following the ‘on’ link of the ‘Advanced Search Options’ in the search interface (Fig. 4). Options include restricting the search field and scope, specifying sort options as well as search modes. Last but not least, searches can be further filtered according to author, journal, year and document ID. Textpresso currently indexes five (bibliographical) fields, abstract, body of text, title, author and year. By default, abstract, body and title are searched, but sometimes it is useful to explicitly choose search fields, which can be done through the corresponding click boxes.
One of the strengths of Textpresso consists of searching through every single sentence and requiring that all query items are met within one sentence. However, the user can also choose to match keywords and categories in search fields only or in the whole document. In the latter case the search behavior is equivalent to that of Google or Yahoo. It is controlled through the option ‘Search Scope’; its default scope is ‘sentence’.
Two other important options are the sort function and additional filtering. The result pages can be sorted according to score (roughly the number of matches in a document, this is the default behavior) or alphabetically according bibliographical fields such as author, year, etc. Lastly, there are two ways of filtering available. Either one filters the search results while formulating the query. In this mode the fields author, journal, year and document ID are available, and any string specified will be partially or completely matched in the respective fields. As an alternative, one can first perform a search, and subsequently the results can be narrowed through a text field. This text field only appears after the initial search has been completed and a search result has been completed. The syntax for this filtering is similar to the PubMed syntax, and is explained in detail on the website.
A second way of using Textpresso for Neuroscience is accomplished via web services. They are implemented as a two-step process. The first step is to run a search on the server; the second step is to retrieve results from the server. This separation of processes is necessary because the search results (in XML format) may be on the order of several megabytes, and forming the XML file may take more time than the time out limit for the client process. In addition, the user may not need all the documents that the search produces. In most cases, users are interested only in the documents (and the sentences therein) that have the maximum scores, similar to how users look only through the first few pages of a Google search. The current set-up allows the client to retrieve a maximum of 500 documents in one call. For retrieving more than 500 documents, the client needs to send more queries with appropriate document numbers.
The web service descriptive language (WSDL) document for the search webservice for Textpresso for Neuroscience is located at http://www.textpresso.org/neuroscience/webservice/wsdl/search.wsdl and is displayed in its current version in Fig. 5. It is used for issuing a query and performing a search on the Textpresso database. The client request sends parameters such as keywords and phrases, categories, search fields and specifications such as ‘exact match’ and ‘case sensitive’ to the server. Single keywords or phrases or combinations of keywords and/or phrases can be submitted, and the Boolean expressions AND, NOT and OR are also allowed. Comma-separated list of categories can also be specified, and they are concatenated by a Boolean ‘AND’ operation to other categories and keywords if present. Currently, Textpresso offers searches in abstract, body and title, which are called search fields. All or a subset of these fields can be entered.
Once the server receives the client request, it performs the search and stores the results in a temporary file. The response from server is a single string of the form ‘SearchID TotalDoc’. The SearchID represents a filename in which the server has stored the results locally and TotalDoc is the total number of documents the search yielded. The search results can then be retrieved in XML format by using the web service described at http://www.textpresso.org/neuroscience/webservice/wsdl/retrieve.wsdl and shown in Fig. 6. The client call sends the parameters SearchID, FirstDoc, LastDoc and a Boolean flag MatchingSentence. SearchID is the identification number for the search performed previously with the search web service (and returned with it). FirstDoc and LastDoc are some positive integers that determine which documents within the range of TotalDoc, the total number of documents of the search, are returned. As the search results on the server are represented as a list sorted by score, FirstDoc is the position of the first document in that list to be retrieved. LastDoc is the position of the last documents to be obtained; it has to be greater than FirstDoc and less than TotalDoc. There is also an additional restriction that LastDoc should not be greater than FirstDoc by more than 500. This ensures that the server returns only a maximum of 500 documents at once. If more than 500 documents are requested at once, the server returns only the first 500 documents. The Boolean flag MatchingSentence determines whether matching sentences should also be returned. If not, only the bibliographies of each result entry are returned. The client is returned a single string, which contains the search results in the form of an XML document.
Textpresso in the Context of the Neuroscience Information Framework
Textpresso is one of the core resources of the Neuroscience Information Framework NIF (Gupta et al., 2008). The NIF search interfaces offer direct querying of Textpresso for Neuroscience through the web service described above. NIF searches can be accessed through http://nif.nih.gov by following the ‘Search NIF’ link. It offers two search modes, a simple and an advanced search. In the simple search the user enters a keyword, and a search is immediately initiated. The advanced search requires entering a term and then picking matching terms of NIF concepts. These terms are then expanded, and the user can opt to add synonyms for her search resulting in a set of terms. Both NIF websites query several resources simultaneously with these terms, and their results can be viewed by clicking on one of the respective tabs. One of the tabs, called ‘Literature,’ displays the results of the query that has been submitted to Textpresso. The results show journal, title, author and year of publication information for each entry, and also links to Textpresso for Neuroscience, PubMed and Google Scholar. Following the Textpresso links allows the user to see the actual matches of the search in the full text, and follow-up searches can be performed at the Textpresso site as the original query from the NIF site is carried through to the Textpresso site.
Conclusion
Textpresso is a powerful tool for the neuroscientist due to its ability to query the full texts of tens of thousands of articles and abstracts as well as its capacity to include semantic concepts in searches. We are planning to expand the corpus to several hundred thousand full text research papers and are currently researching how scaling the corpus to this size will affect the performance of the system. In addition, the large corpus can be subdivided according to research themes, and the sub-corpora should be made available separately for searching to gain even more specificity. We have previously developed document classification algorithms (Chen et al. 2006) that can easily be applied to this task. Finally, we would like to explore the opportunity to interact with the NIF interface via NIF concepts. NIF currently queries Textpresso via a set of terms concatenated with Boolean OR or AND, which becomes unfeasible when several dozen terms are included. Concept-based queries are much more efficient and a natural way of querying. The NIF interface would then query Textpresso by passing a NIF concept ID. This ID is mapped to a corresponding Textpresso category whose lexicon has been filled with NIF vocabularies beforehand. A NIF concept search then simply becomes a one-category search for Textpresso.
Information Sharing Statement
The Textpresso for Neuroscience site is available at http://www.textpresso.org/neuroscience and through the NIF website at http://nif.nih.gov. The software can be downloaded from the Textpresso homepage at http://www.textpresso.org/. Any results and output obtained from the TextpressoTM server shall not be used for any purpose other than private study, scholarship or academic research. Anybody using Textpresso in excess of "fair use" may be liable for copyright infringement. Further online resources mentioned in this article are:
-
Google. http://www.google.com/.
-
Google Scholar. http://scholar.google.com/.
-
GoPubMed. http://www.gopubmed.org.
-
NCBI, National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov/.
-
NIDA, National Institute on Drug Abuse. http://www.drugabuse.gov/.
-
NIF. http://nif.nih.gov/.
-
PubMed Central. http://www.pubmedcentral.nih.gov/.
-
PubMed E-utilities. http://eutils.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html.
-
Textpresso (main site). http://www.textpresso.org/.
-
Textpresso for Neuroscience. http://www.textpresso.org/neuroscience/
Textpresso for Neuroscience search web service. http://www.textpresso.org/neuroscience/webservice/wsdl/search.wsdl
Textpresso for Neuroscience retrieval web service. http://www.textpresso.org/neuroscience/webservice/wsdl/retrieve.wsdl
-
The Gene Ontology. http://www.geneontology.org/.
-
XPDF PDF-to-HTML package. http://pdftohtml.sourceforge.net/.
-
Yahoo. http://www.yahoo.com/.
References
Chen, D., Müller, H.-M., & Sternberg, P. W. (2006). Automatic document classification of biological literature. BMC Bioinformatics, 7, 370. doi:10.1186/1471-2105-7-370.
Doms, A., & Schroeder, M. (2005). GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Research, 33, W783–W786. doi:10.1093/nar/gki470.
Gardner, D., Akil, H., Ascoli, G. A., Bowden, D. M., Bug, W., Donohue, D. E., Goldberg, D. H., Grafstein, B., Grethe, J. S., Gupta, A., Halavi, M., Kennedy, D. N., Marenco, L., Martone, M. E., Miller, P. L., Müller,-H. M., Robert, A., Shepherd, G. M., Sternberg, P. W., Van Essen, D. C., & Williams, R. W. (2008). The Neuroscience Information Framework: a data and knowledge environment for neuroscience. Neuroinformatics, this issue.
Gupta, A., Bug, W., Marenco, L., Qian, X., Condit, C., Rangarajan, A., Müller, H. M., Miller, P. L., Sanders, B., Grethe, J. S., Astakhov, V., Shepherd, G. M., Sternberg, P. W., & Martone, M. E. (2008). Federated access to heterogeneous information resources in the Neuroscience Information Framework (NIF). Neuroinformatics, this issue.
Hunter, L., & Cohen, K. B. (2006). Biomedical language processing: perspective what’s beyond PubMed? Molecular Cell, 21, 589–594. doi:10.1016/j.molcel.2006.02.012.
Müller, H.-M., Kenny, E., & Sternberg, P. W. (2004). Textpresso: an ontology-based information retrieval system for the biological literature. PLoS Biology, 2(11), e309. doi:10.1371/journal.pbio.0020309.
The Gene Ontology Consortium (2000). Gene Ontology: tool for the unification of biology. Nature Genetics, 25, 25–29. doi:10.1038/75556.
The Gene Ontology Consortium (2008). The Gene Ontology project in 2008. Nucleic Acids Research, 36, D440–D441. doi:10.1093/nar/gkm883.
Zweigenbaum, P., Demner-Fushman, D., Yu, H., & Cohen, K. B. (2007). Frontiers of biomedical text mining: current progress. Briefings in Bioinformatics, 8(5), 358–375. doi:10.1093/bib/bbm045.
Acknowledgement
Textpresso is supported by a grant from the National Human Genome Research Institute at the US National Institutes of Health # HG004090. The Textpresso for Neuroscience site has been funded in whole or in part through the NIH Blueprint for Neuroscience Research with Federal funds from the National Institute on Drug Abuse, National Institutes of Health, Department of Health and Human Services, under contract No. HHSN271200577531C. PWS is an Investigator of the Howard Hughes Medical Institute.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Müller, HM., Rangarajan, A., Teal, T.K. et al. Textpresso for Neuroscience: Searching the Full Text of Thousands of Neuroscience Research Papers. Neuroinform 6, 195–204 (2008). https://doi.org/10.1007/s12021-008-9031-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-008-9031-0